# Clone the repository
git clone https://github.com/zhyan0603/GPUMDkit.git
# Set environment variables
export GPUMDkit_path=/your_dir_of_GPUMDkit
export PATH=${GPUMDkit_path}:${PATH}
source ${GPUMDkit_path}/Scripts/utils/completion.sh
# Make script executable and run
cd $GPUMDkit_path
chmod +x gpumdkit.sh
gpumdkit.shGPUMDkit
A Toolkit for GPUMD & NEP
GPUMDkit is a toolkit for the GPUMD (Graphics Processing Units Molecular Dynamics) and NEP (neuroevolution potential) program. It provides a set of tools to streamline the use of common scripts in GPUMD and NEP, simplifying workflows and enhancing efficiency.
Get StartedKey Features
Script Invocation
Easily run scripts for GPUMD and NEP
Workflow Automation
Automate common tasks to save time and reduce manual intervention
User-Friendly Interface
Intuitive shell commands designed to enhance user experience
Analysis Tools
Comprehensive tools for analyzing simulation results
Quick Start
Documentation
Gallery
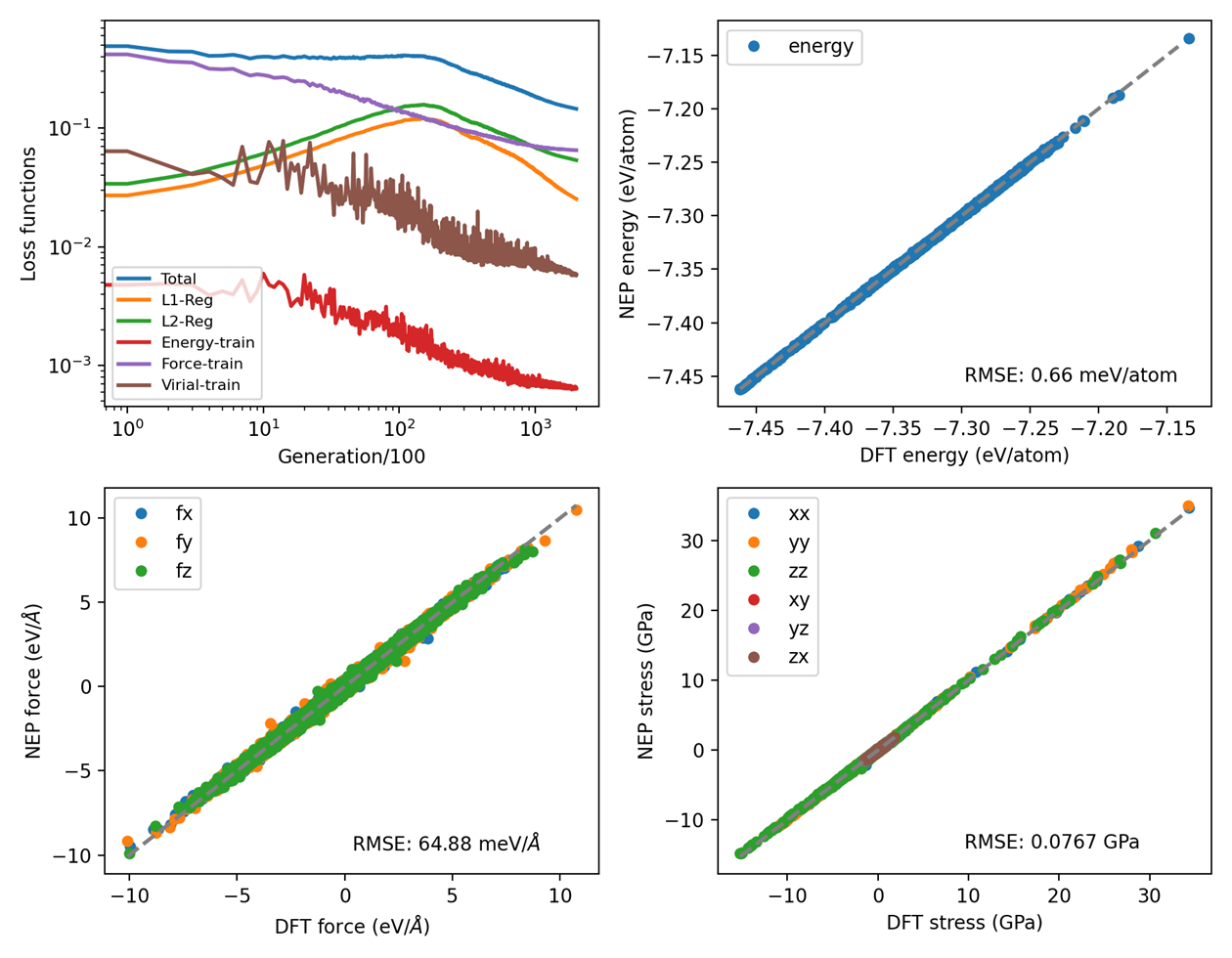
Training Process
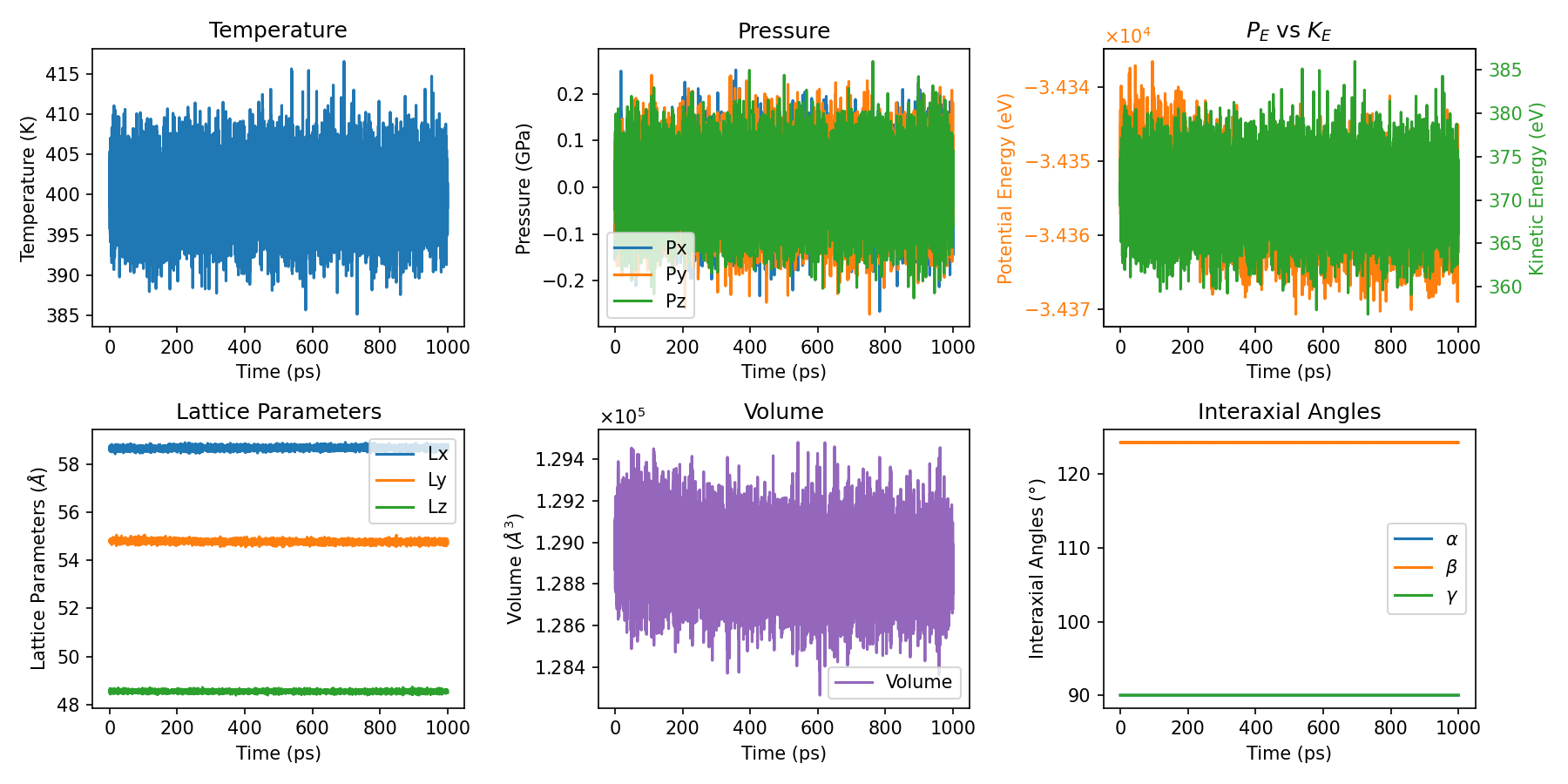
Thermo Analysis
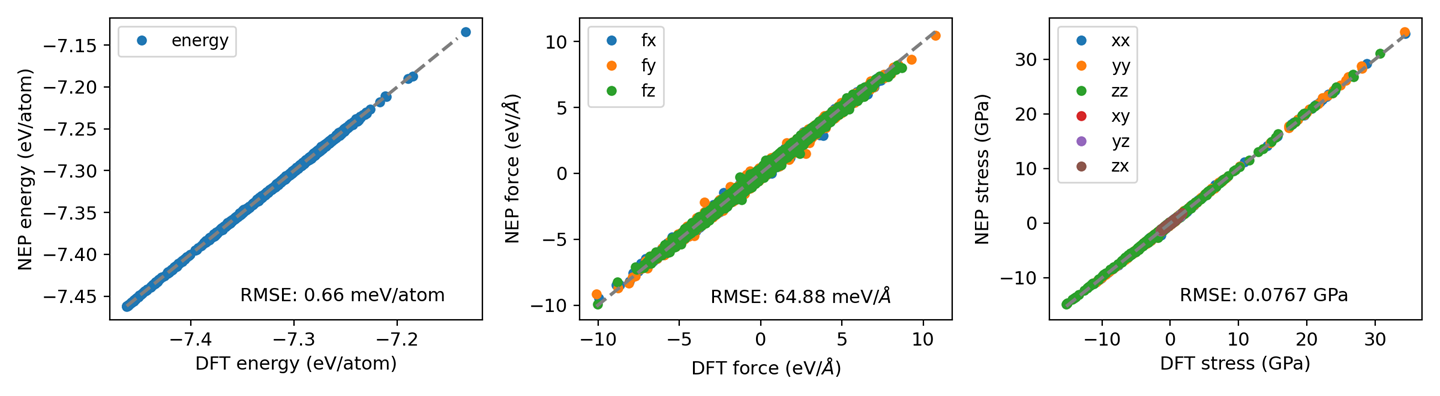
NEP Predictions
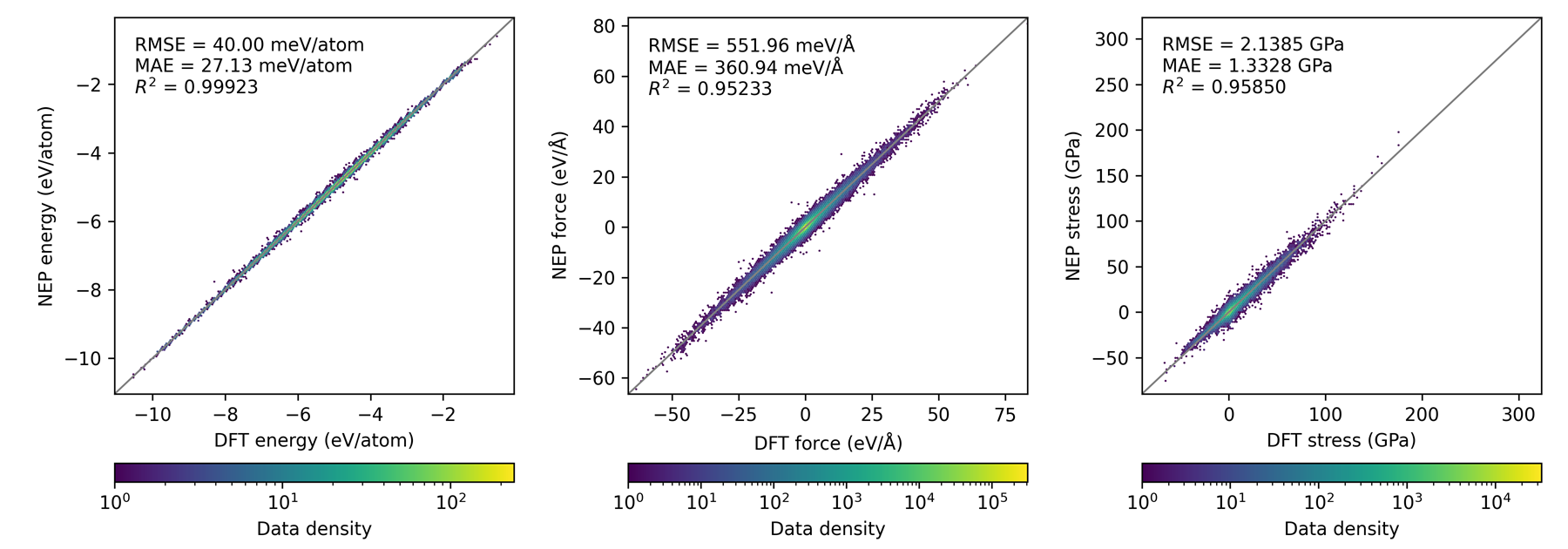
Density Parity Plot
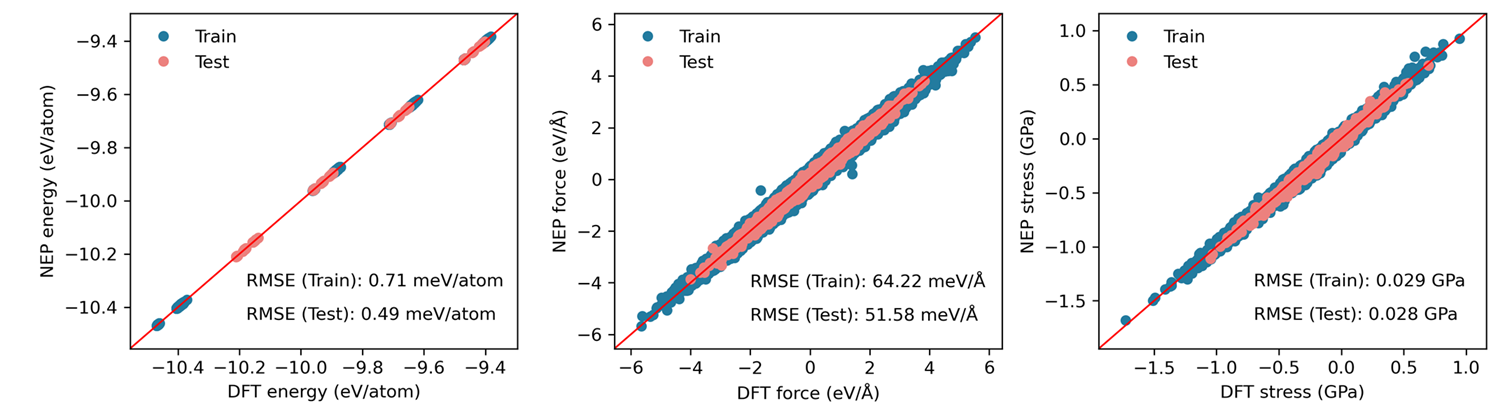
Training and Testing
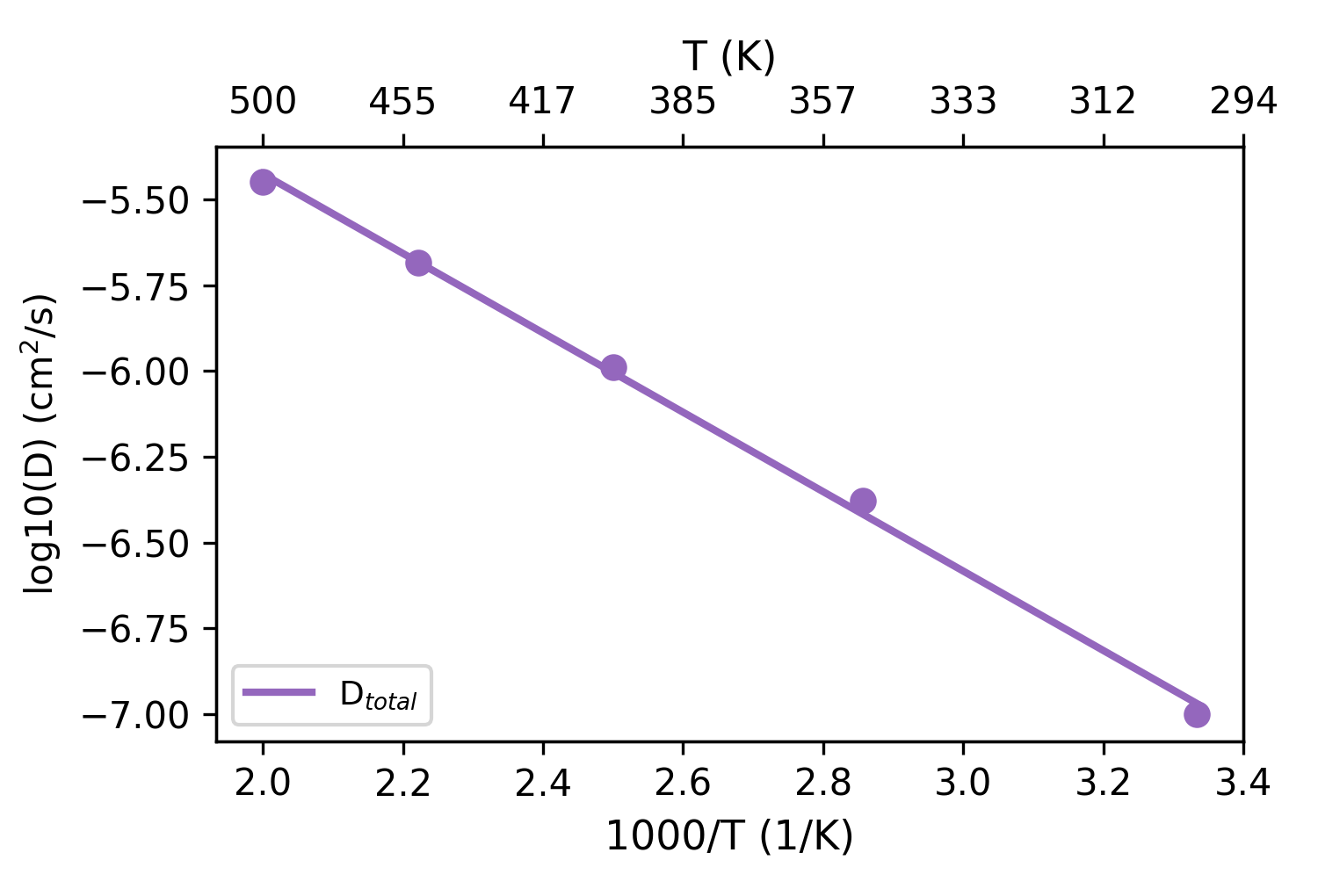
Arrhenius Plot
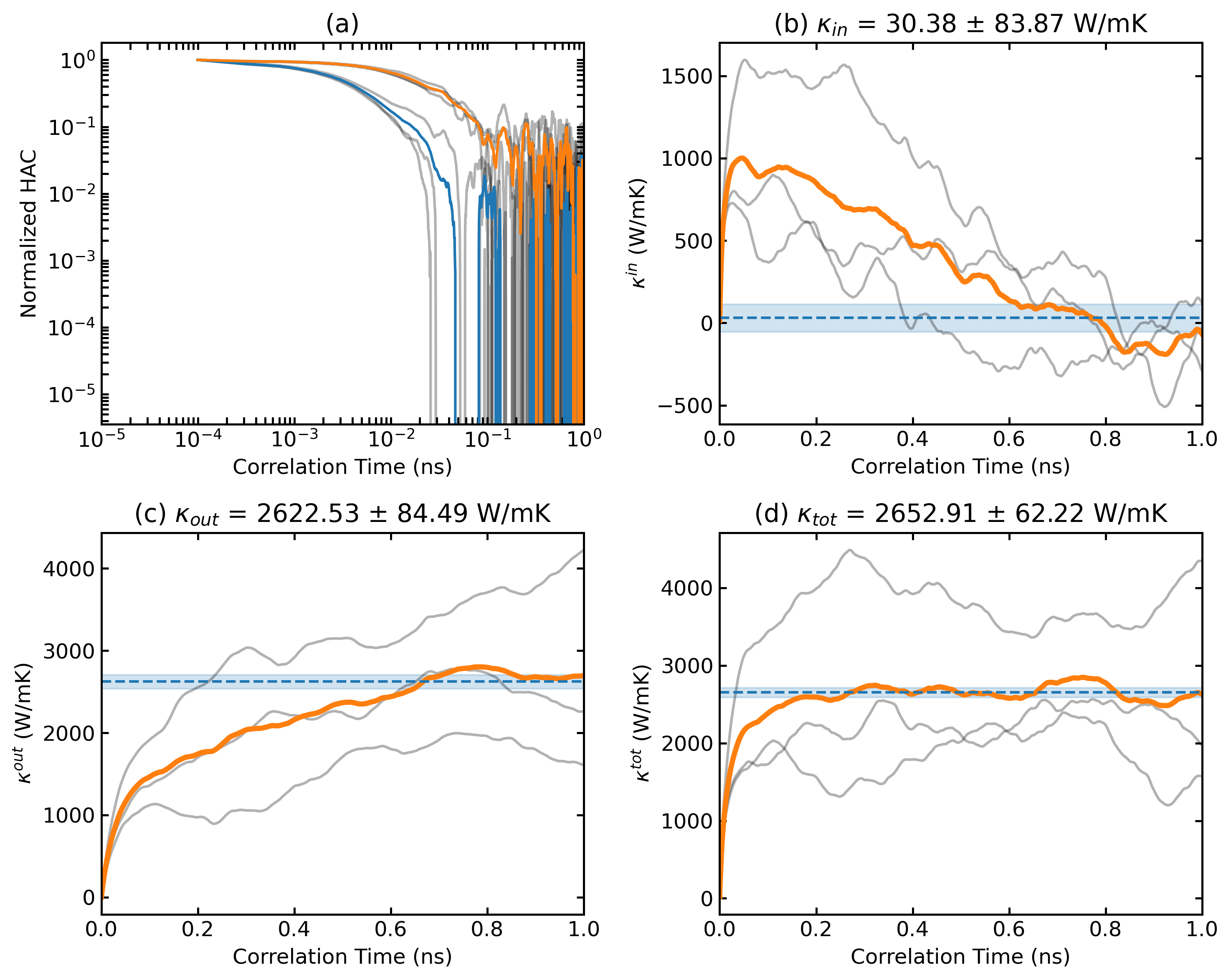
EMD Analysis
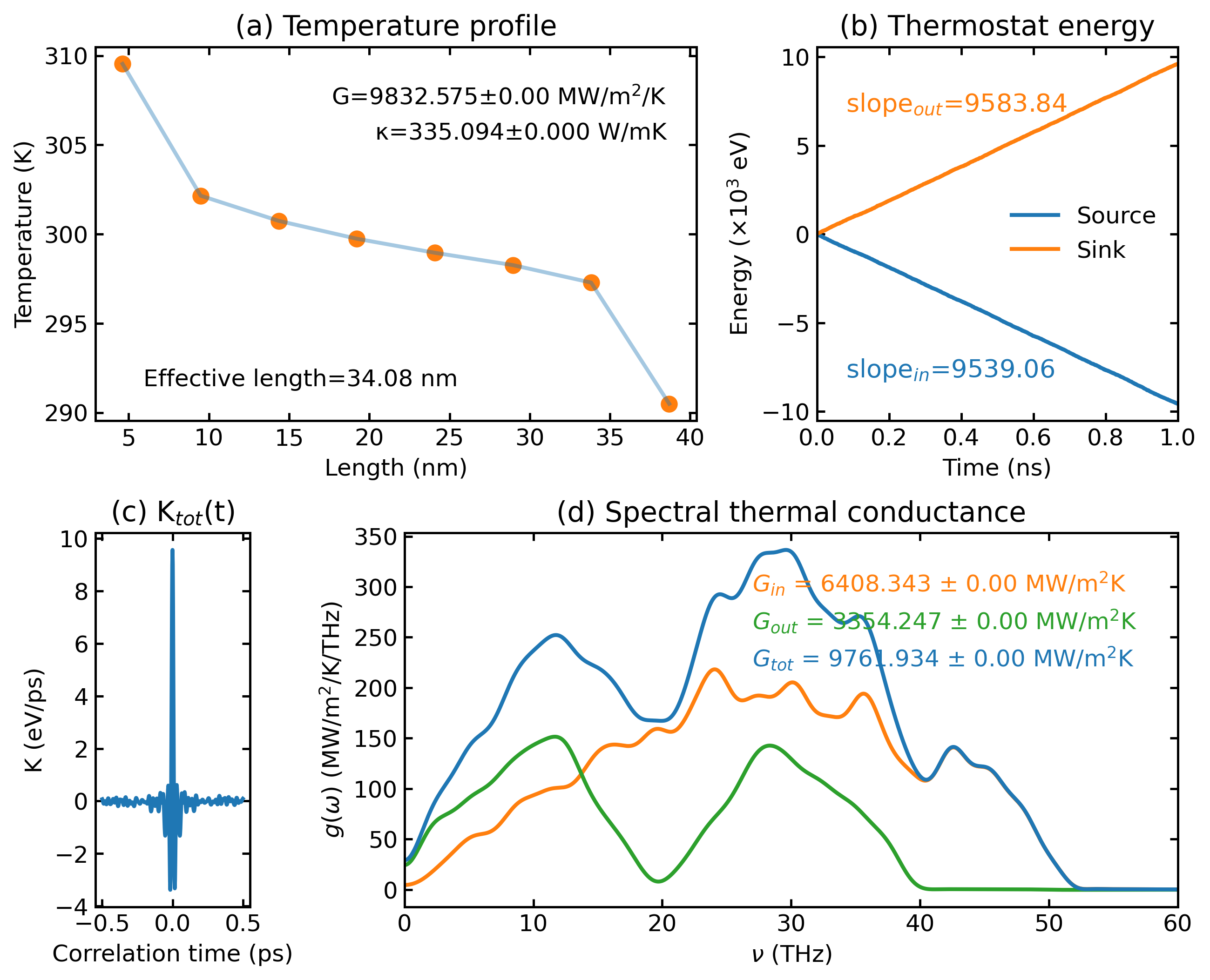
NEMD Analysis
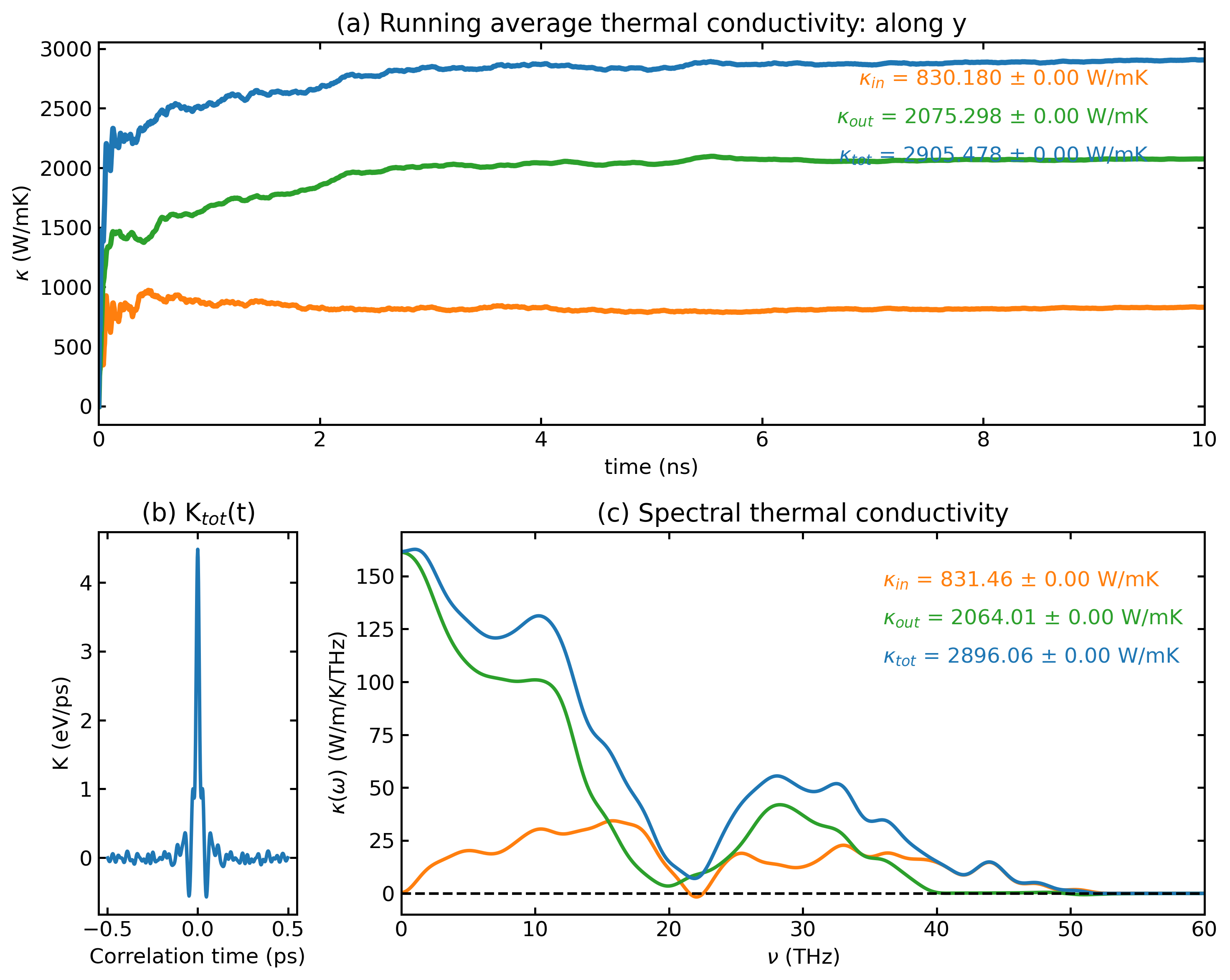
HNEMD Analysis
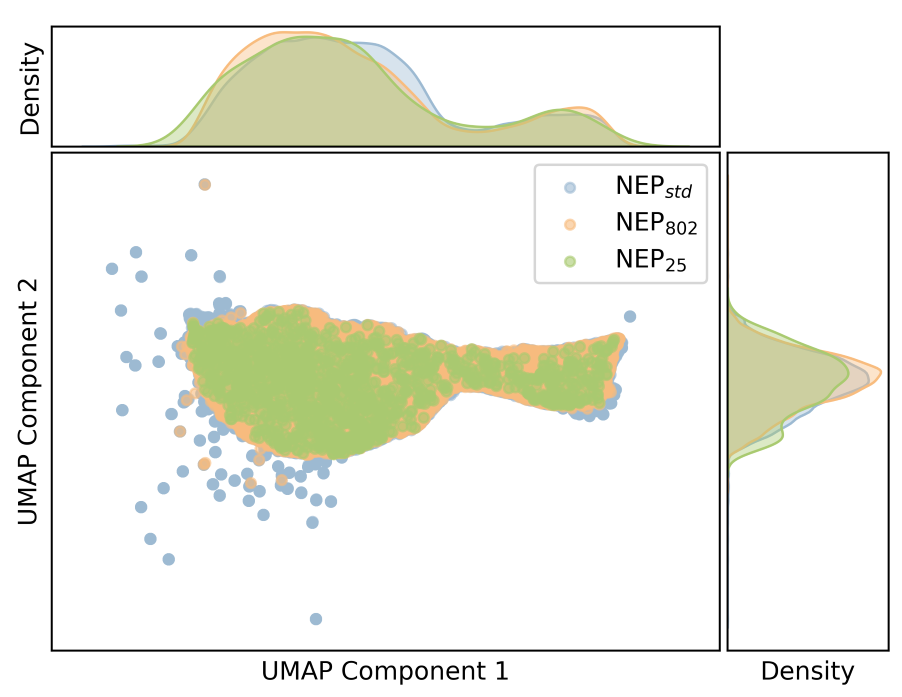
Descriptors Analysis
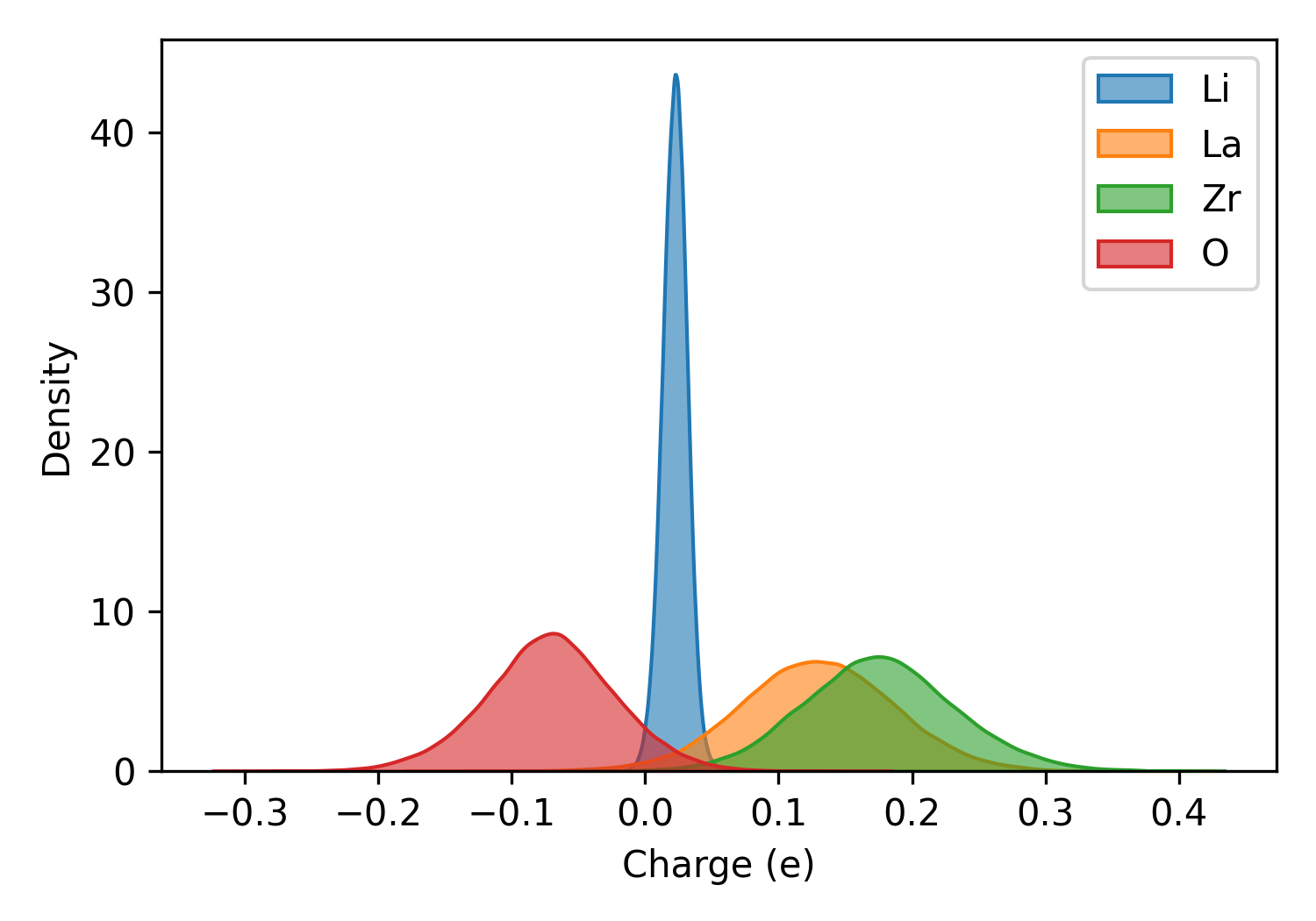
Charge Analysis
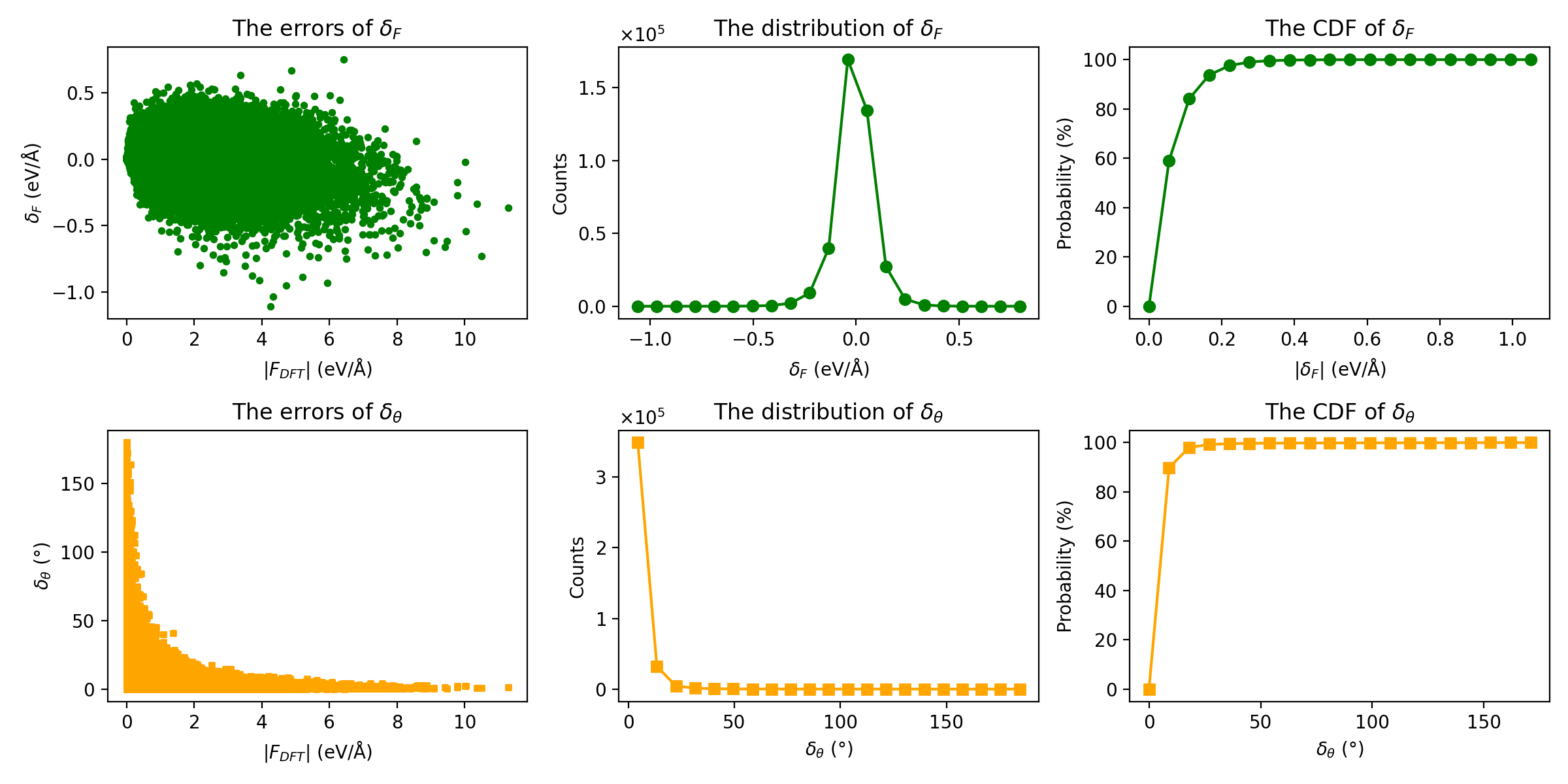
Force Errors
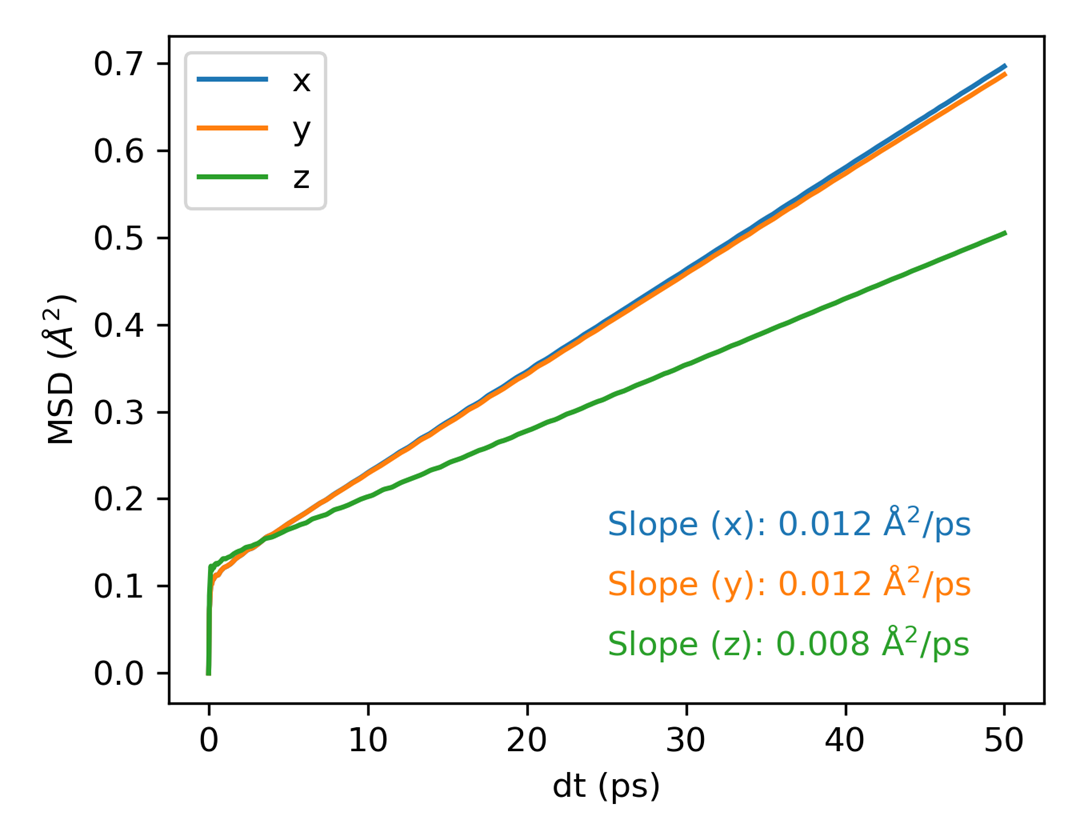
Mean Square Displacement
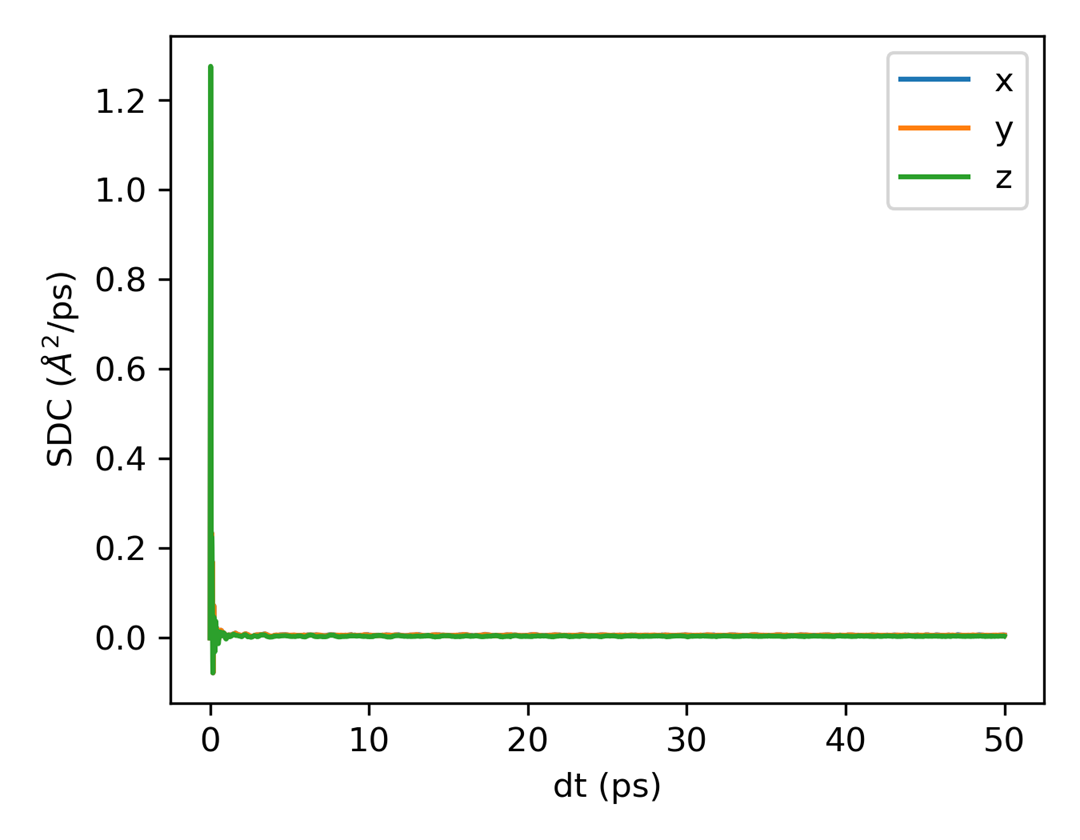
Self Diffusion Coefficient
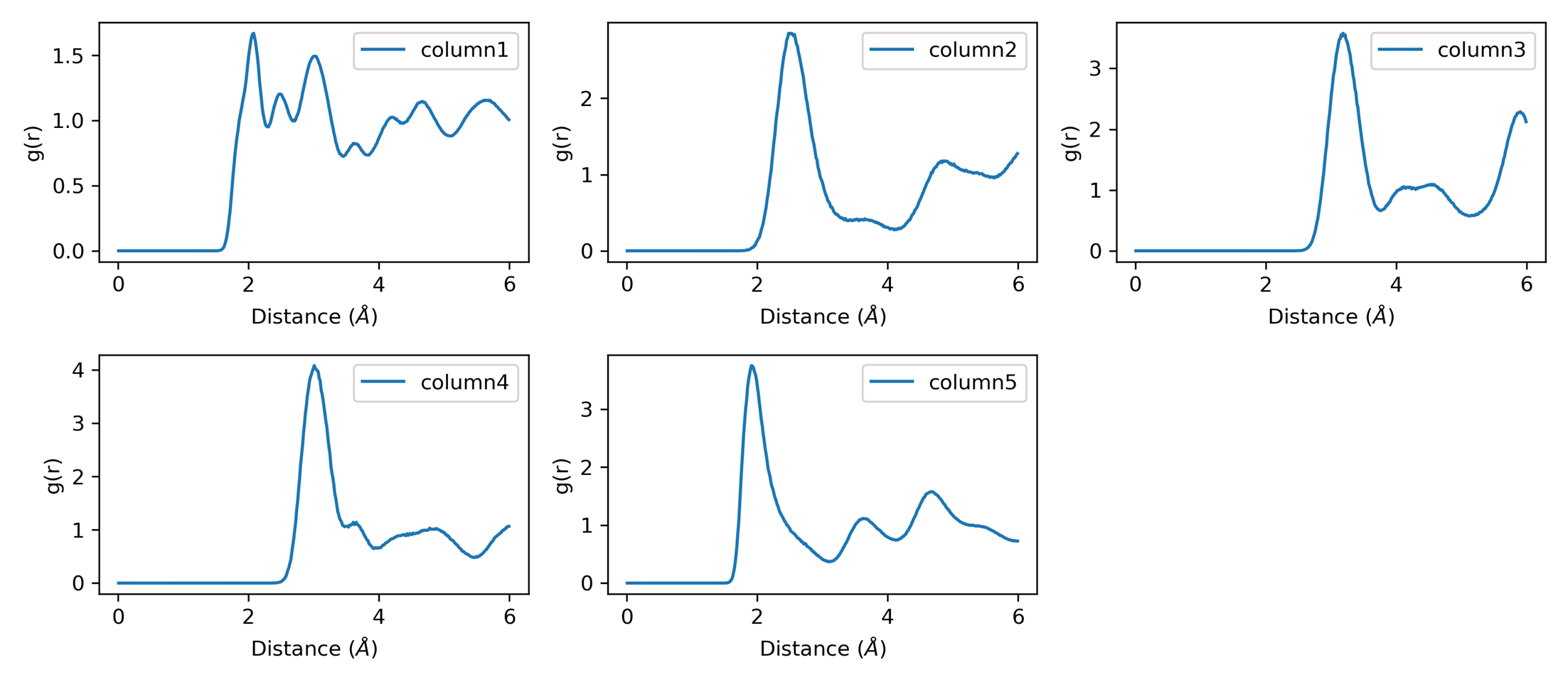
RDF Analysis 1
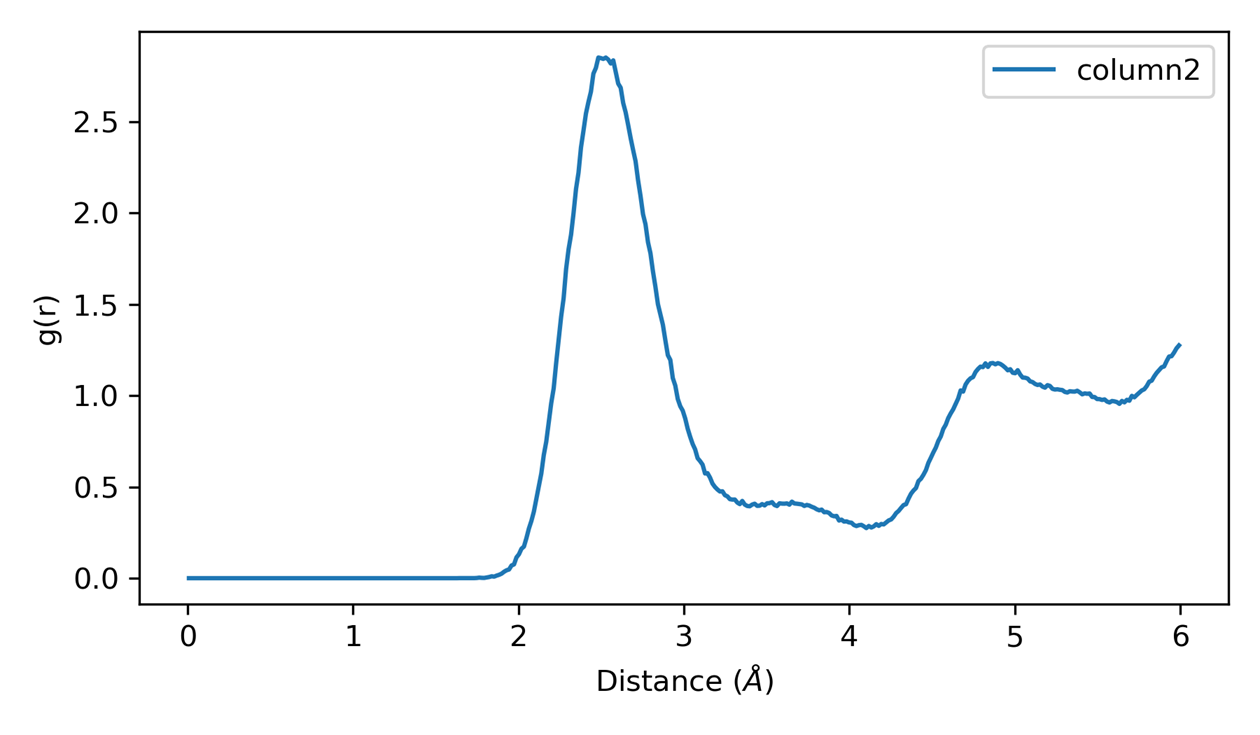
RDF Analysis 2
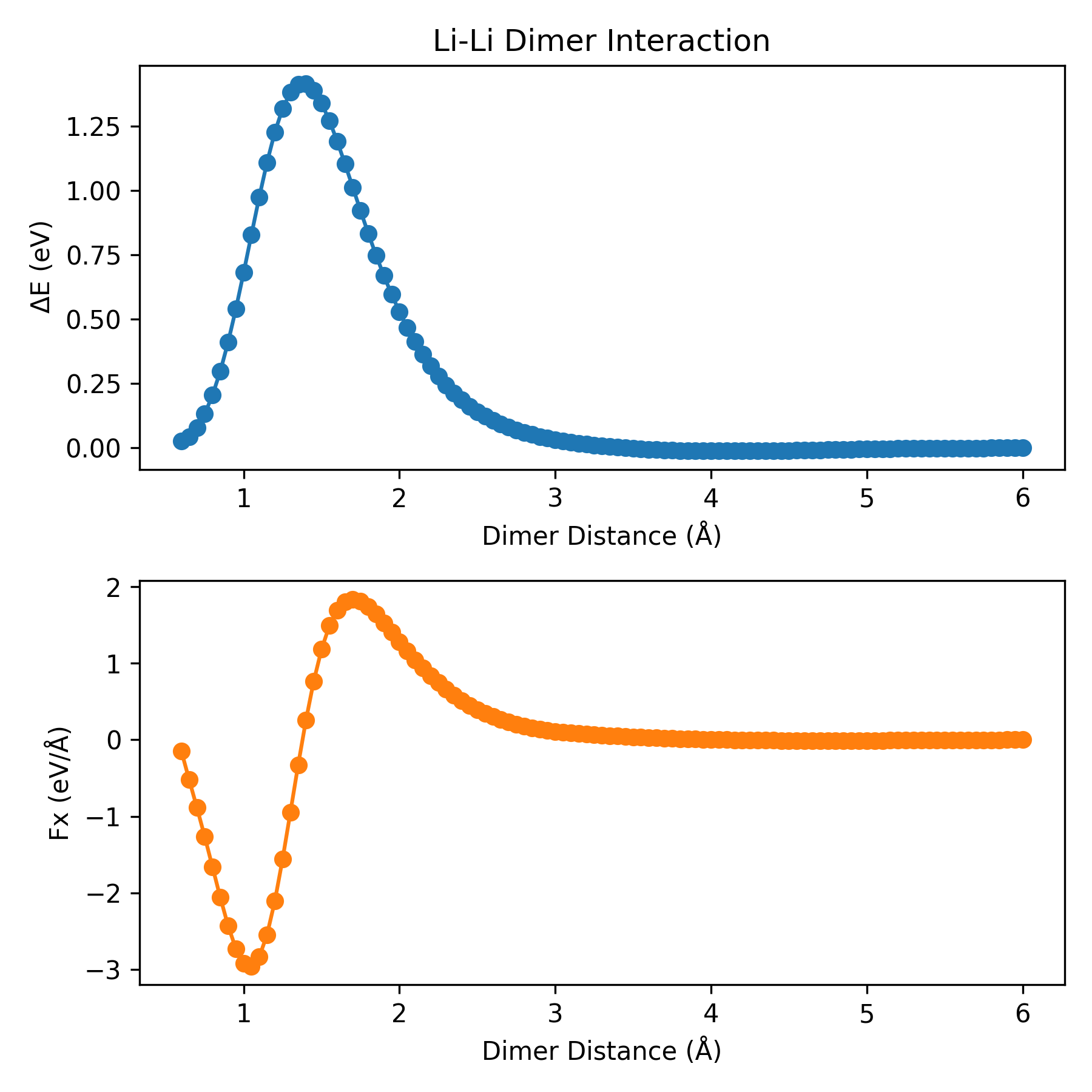
Dimer Plot
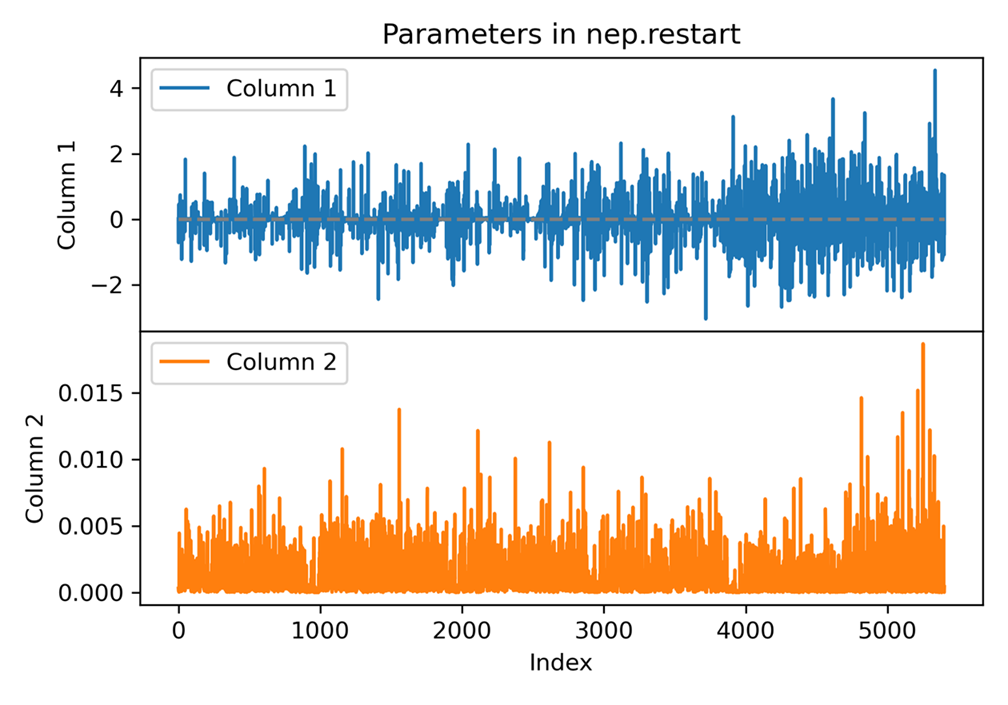
plot nep.restart
Publications
Join Us & Citation
Contribute Code
Help improve GPUMDkit by contributing Python/Shell scripts through Pull Requests.
Submit PRCitation
As of now, our article is still in preparation. If you like it, please star us on GitHub.
Star on GitHub