📊 Plot Scripts
This directory (Scripts/plt_scripts/) contains Python scripts for comprehensive visualization of data generated by GPUMD and NEP. These plotting tools help analyze molecular dynamics simulations, NEP training results, and various material properties.
Script Location: Scripts/plt_scripts/
GPUMDkit provides comprehensive visualization tools for GPUMD and NEP data.
Command-Line Mode
For a complete list:
gpumdkit.sh -plt -h
+=====================================================================================================+
| GPUMDkit 1.4.2 (dev) (2025-12-17) Plotting Usage |
+=============================================== Plot Types ==========================================+
| thermo Plot thermo info | train Plot NEP train results |
| prediction Plot NEP prediction results | train_test Plot NEP train and test results |
| msd Plot mean square displacement | msd_conv Plot the convergence of MSD |
| msd_all Plot MSD of all species | sdc Plot self diffusion coefficient |
| rdf Plot radial distribution function | vac Plot velocity autocorrelation |
| restart Plot parameters in nep.restart | dimer Plot dimer plot |
| force_errors Plot force errors | des Plot descriptors |
| charge Plot charge distribution | lr Plot learning rate |
| doas Plot density of atomistic states | arrhenius_d Plot Arrhenius diffusivity |
| arrhenius_sigma Plot Arrhenius sigma | net_force Plot net force distribution |
| emd Plot EMD results | nemd Plot NEMD results |
| hnemd Plot HNEMD results | |
+=====================================================================================================+
| For detailed usage and examples, use: gpumdkit.sh -plt <plot_type> -h |
+=====================================================================================================+
See the codes in plt_scripts for more details
Code path: /d/Westlake/GPUMD/Gpumdkit/Scripts/plt_scripts
Script Categories
🔬 NEP Training & Prediction
Scripts for monitoring and analyzing NEP model training and predictions.
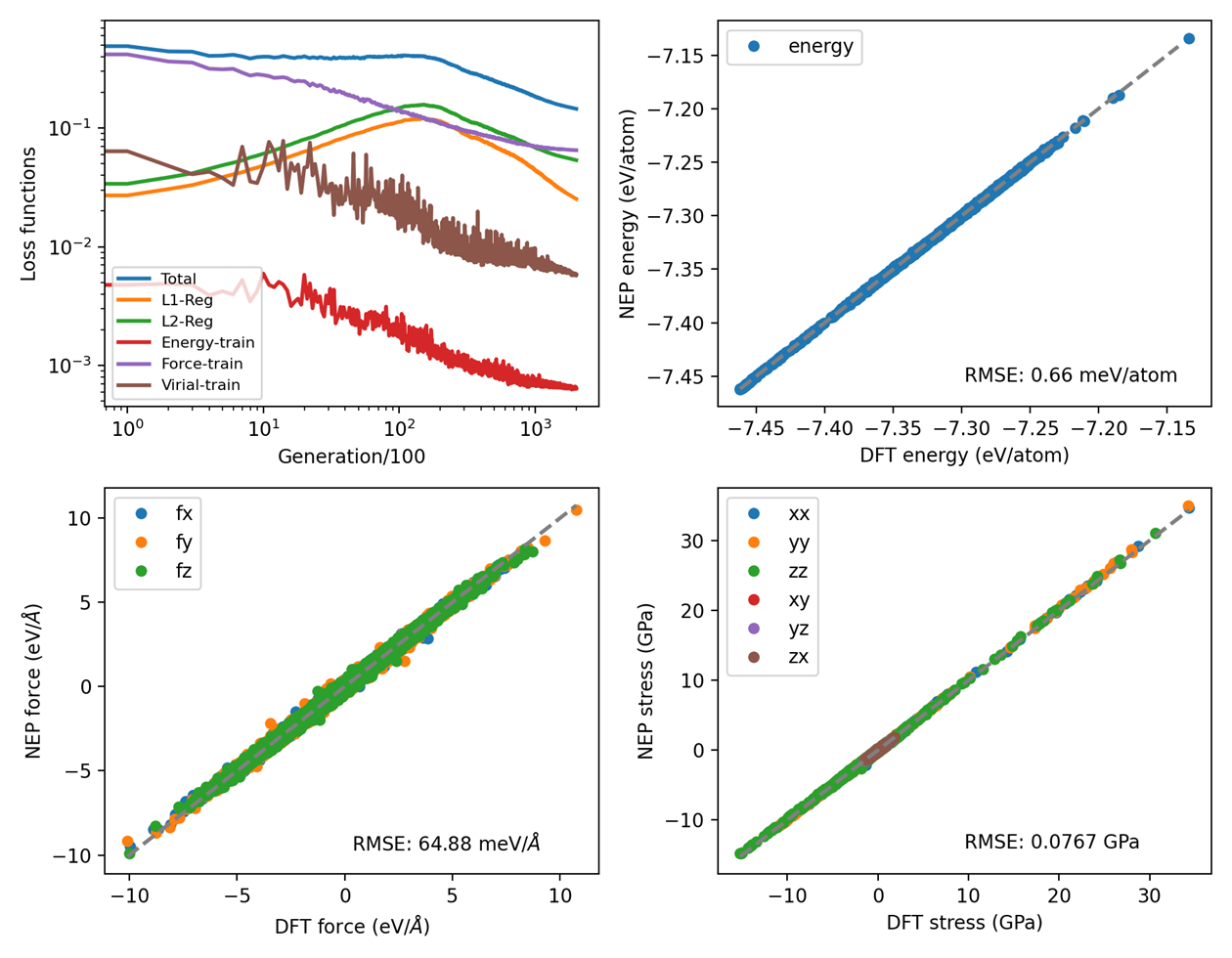
plt_train.py
Visualizes NEP training progress including loss curves, RMSE evolution, and parity plots comparing DFT vs NEP predictions for energy, forces, and stresses.
Input File: loss.out, energy_train.out, force_train.out, virial_train.out
Usage:
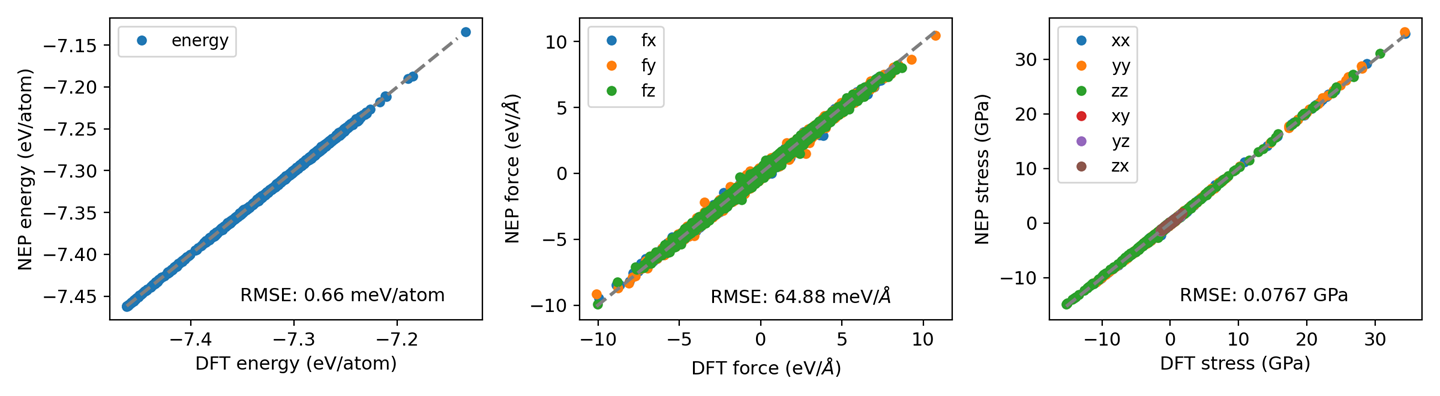
plt_prediction.py
Visualizes NEP prediction results.
Input Files: energy_test.out, force_test.out, virial_test.out
Usage:
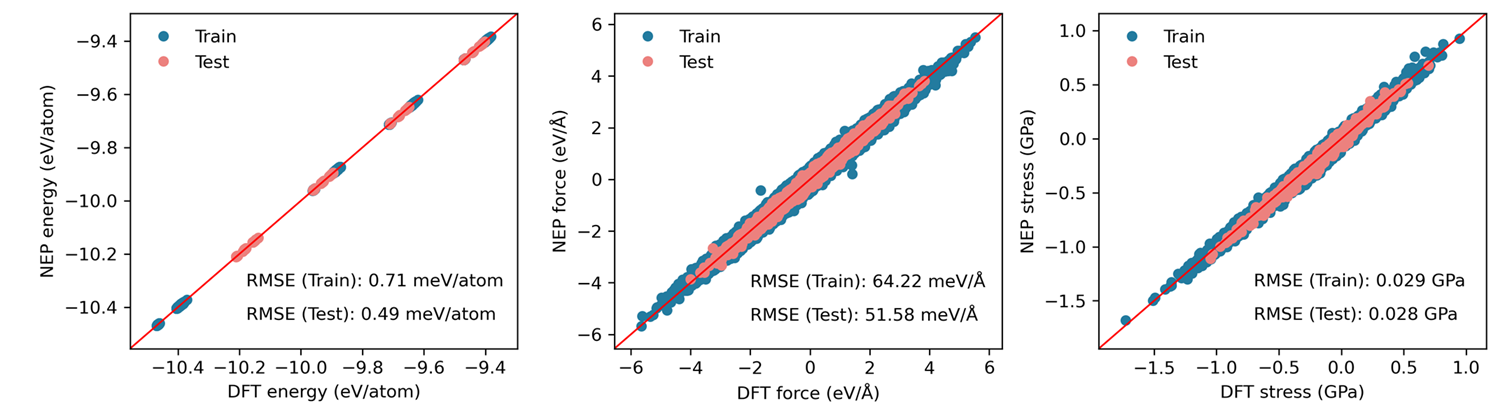
plt_train_test.py
Creates combined parity plots for both training and testing datasets.
Input Files: *_train.out, *_test.out
Usage:
plt_parity_density.py
Generates density-based parity plots for energies, forces, and stresses, useful for large datasets.
Input Files: energy_train.out, force_train.out, virial_train.out
Usage:

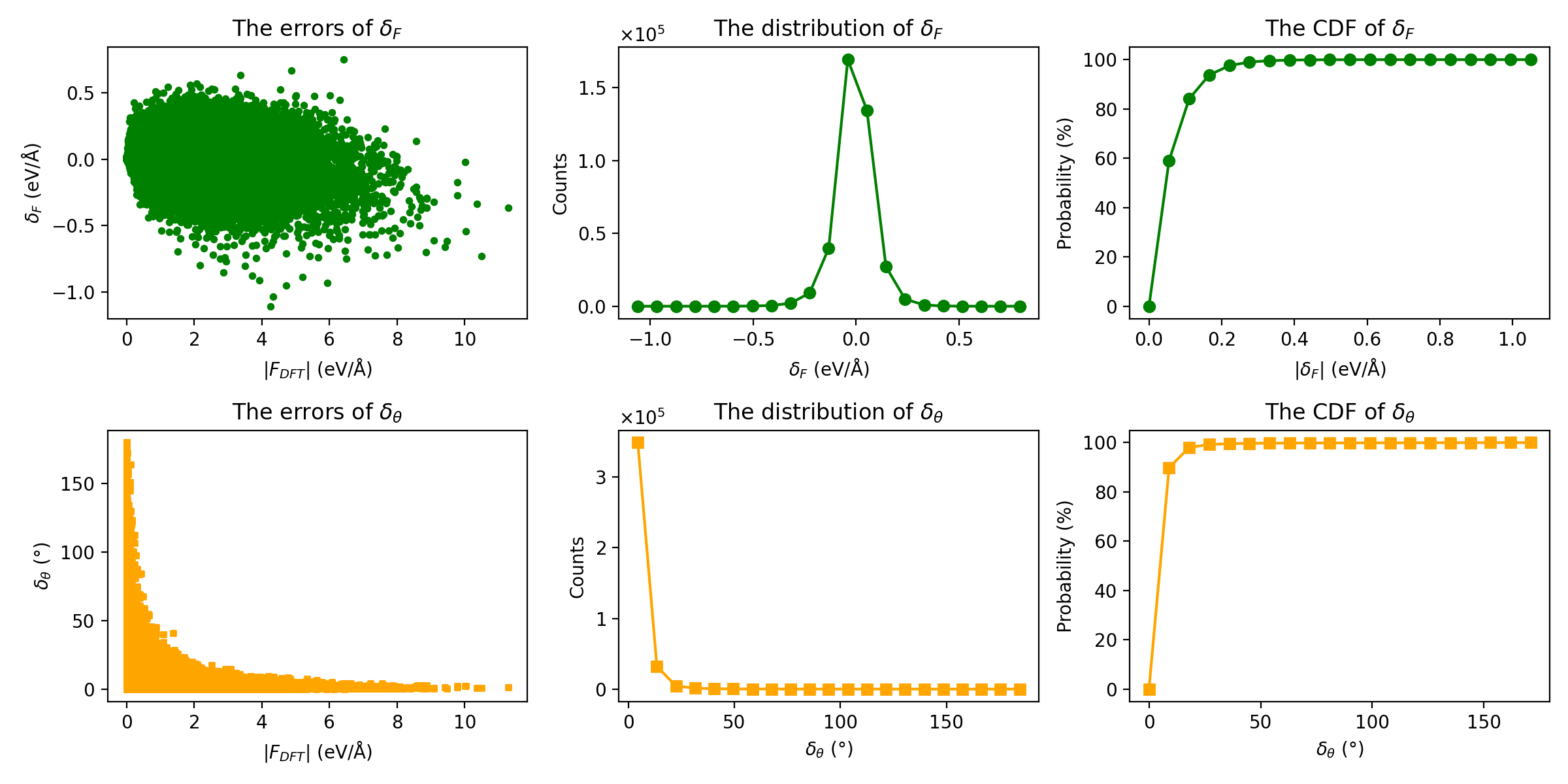
plt_force_errors.py
Plots force error evaluation metrics as proposed in Liu et al..
Input File: force_train.out
Usage:
Metrics Displayed: - Force magnitude errors (delta_F) - Force angle errors (delta_theta) - Distribution of errors
plt_learning_rate.py
Visualizes learning rate during gnep training.
Input File: loss.out
Usage:
Use Case: Monitor learning rate decay.
Note: this is only for the gnep training process.
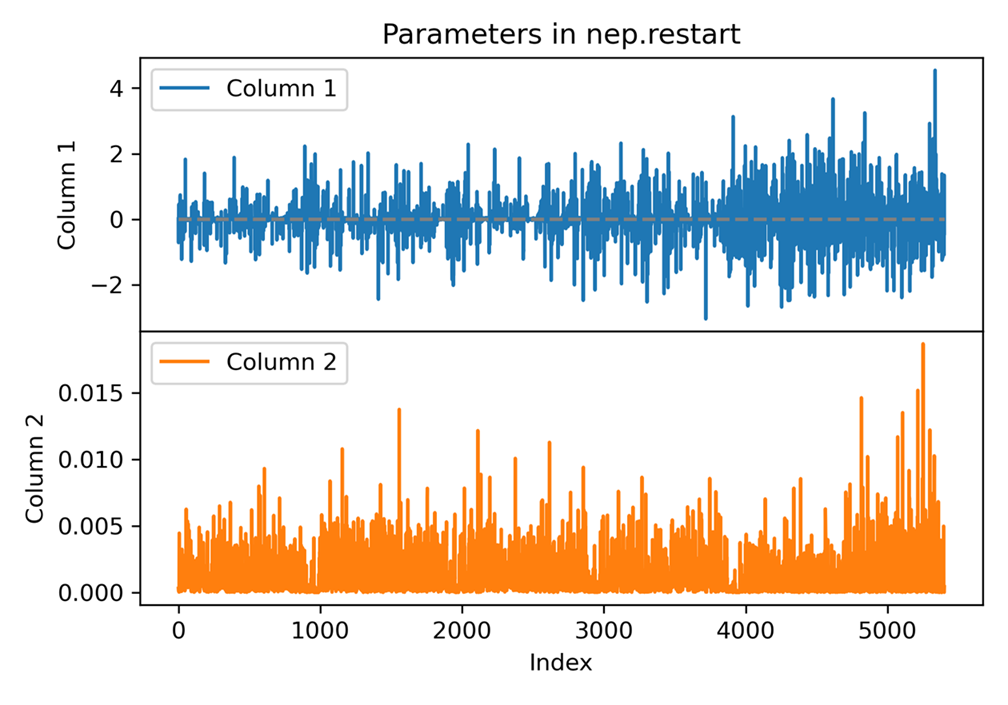
plt_nep_restart.py
Visualizes parameters stored in nep.restart file.
Input File: nep.restart
Usage:
🌡️ Thermodynamic Properties
Scripts for visualizing thermo info from GPUMD simulations.
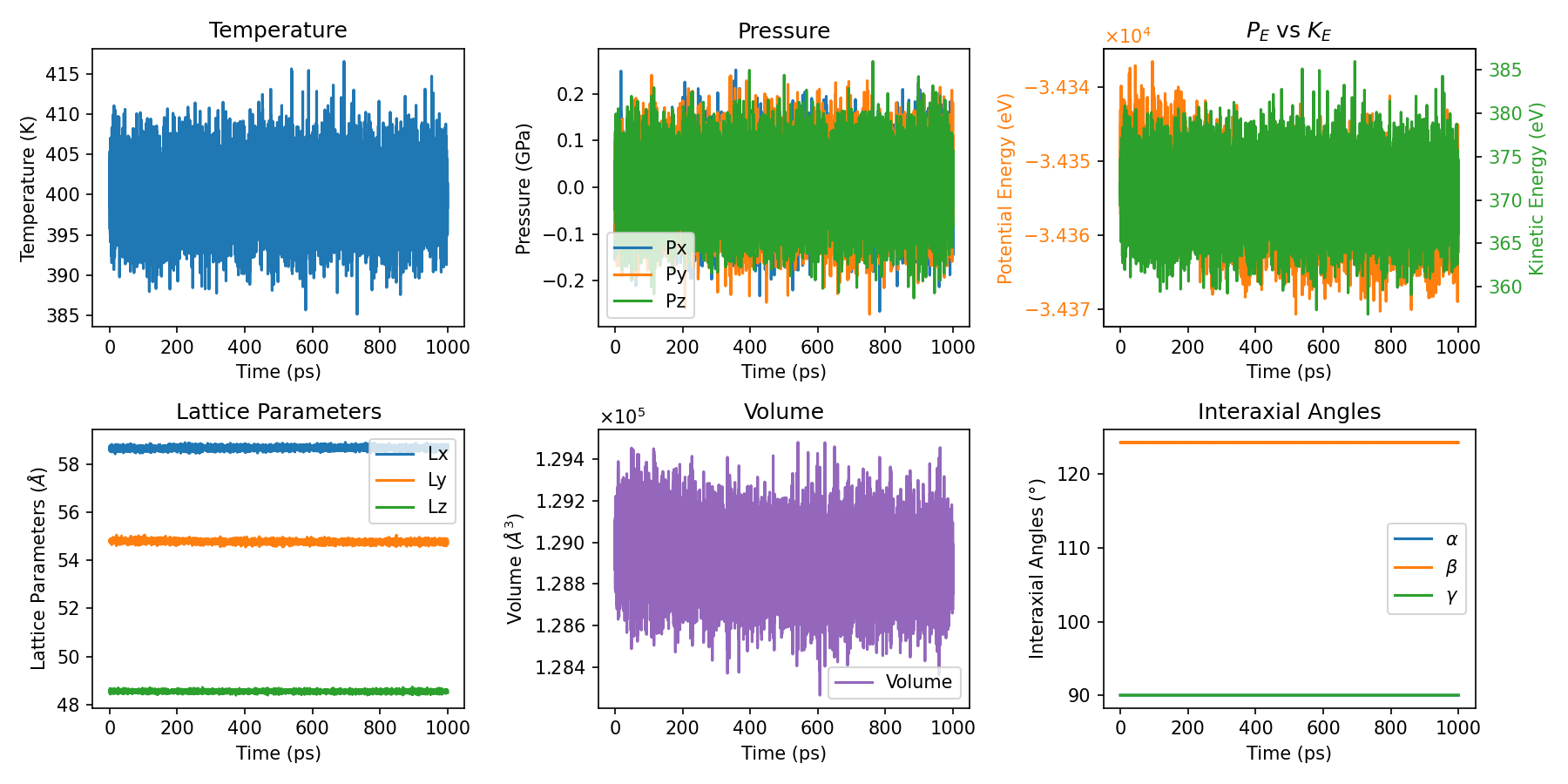
plt_thermo.py
Primary script for comprehensive thermodynamic property visualization.
Input File: thermo.out
Usage:
plt_thermo2.py & plt_thermo3.py
Alternative thermodynamic visualization (different styles).
Usage:
🔄 Transport Properties
Scripts for analyzing diffusion, ionic conductivity, and thermal transport, etc.
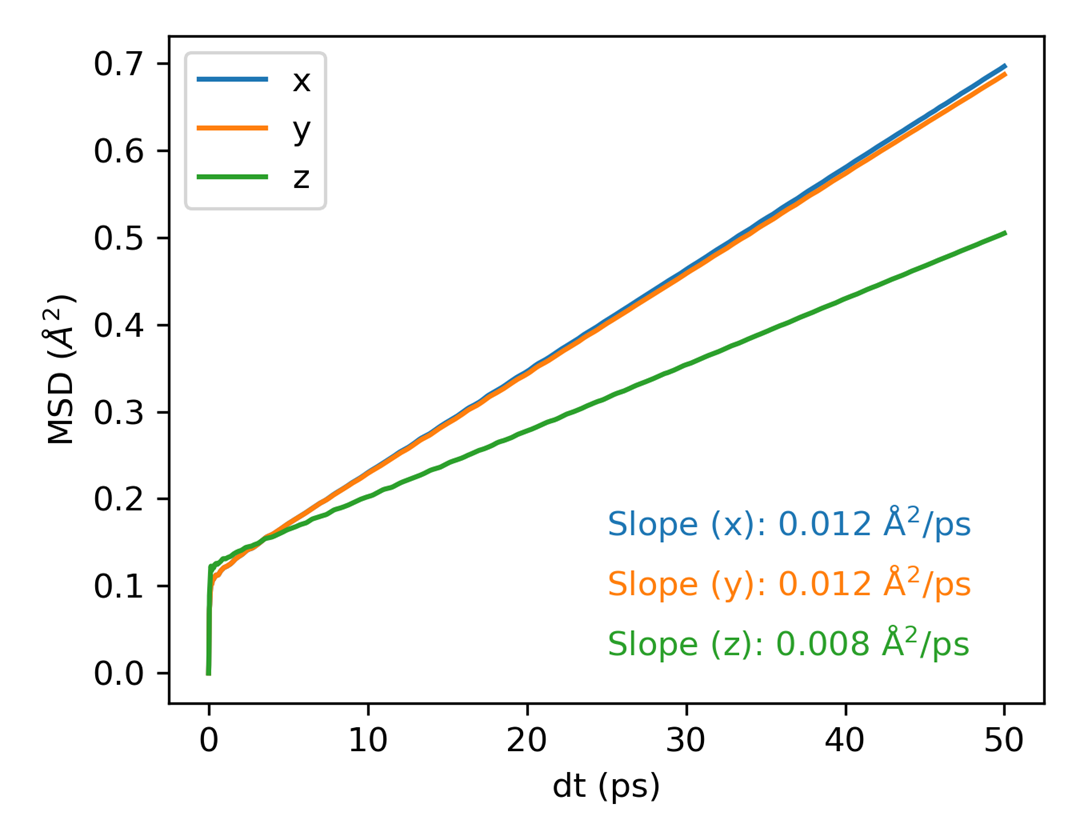
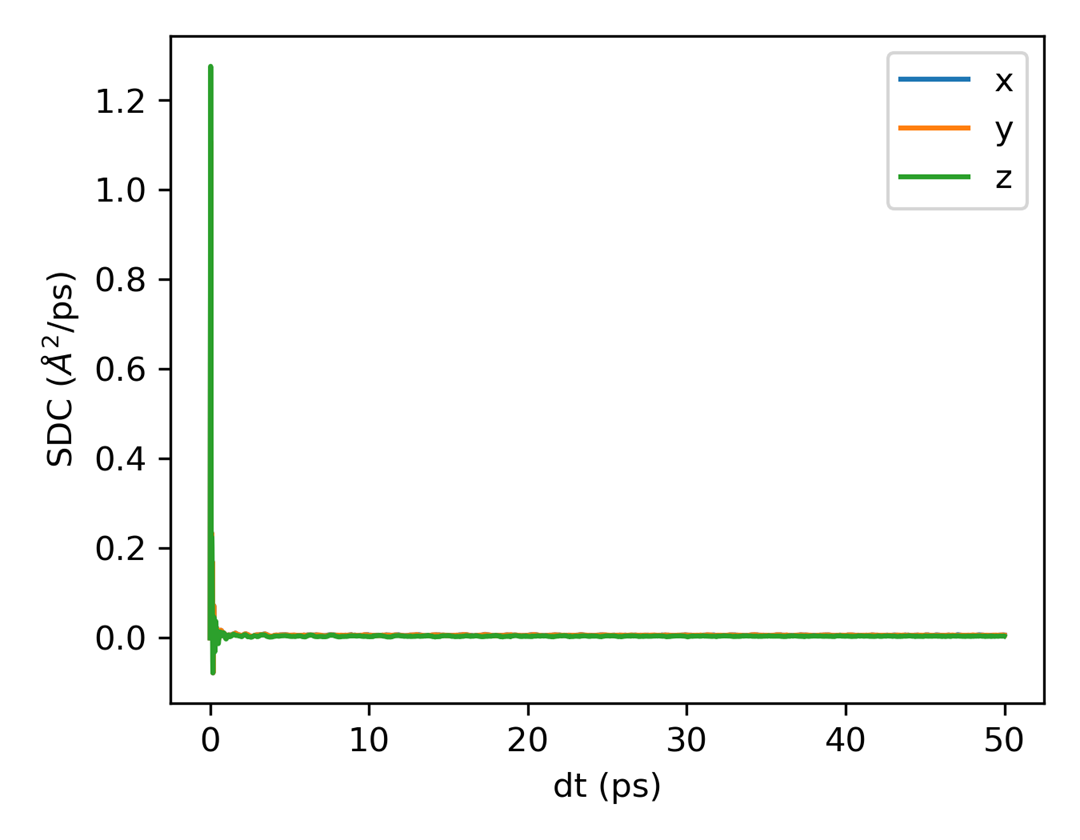
plt_msd.py
Plots mean square displacement (MSD) for all directions.
Input File: msd.out
Usage:
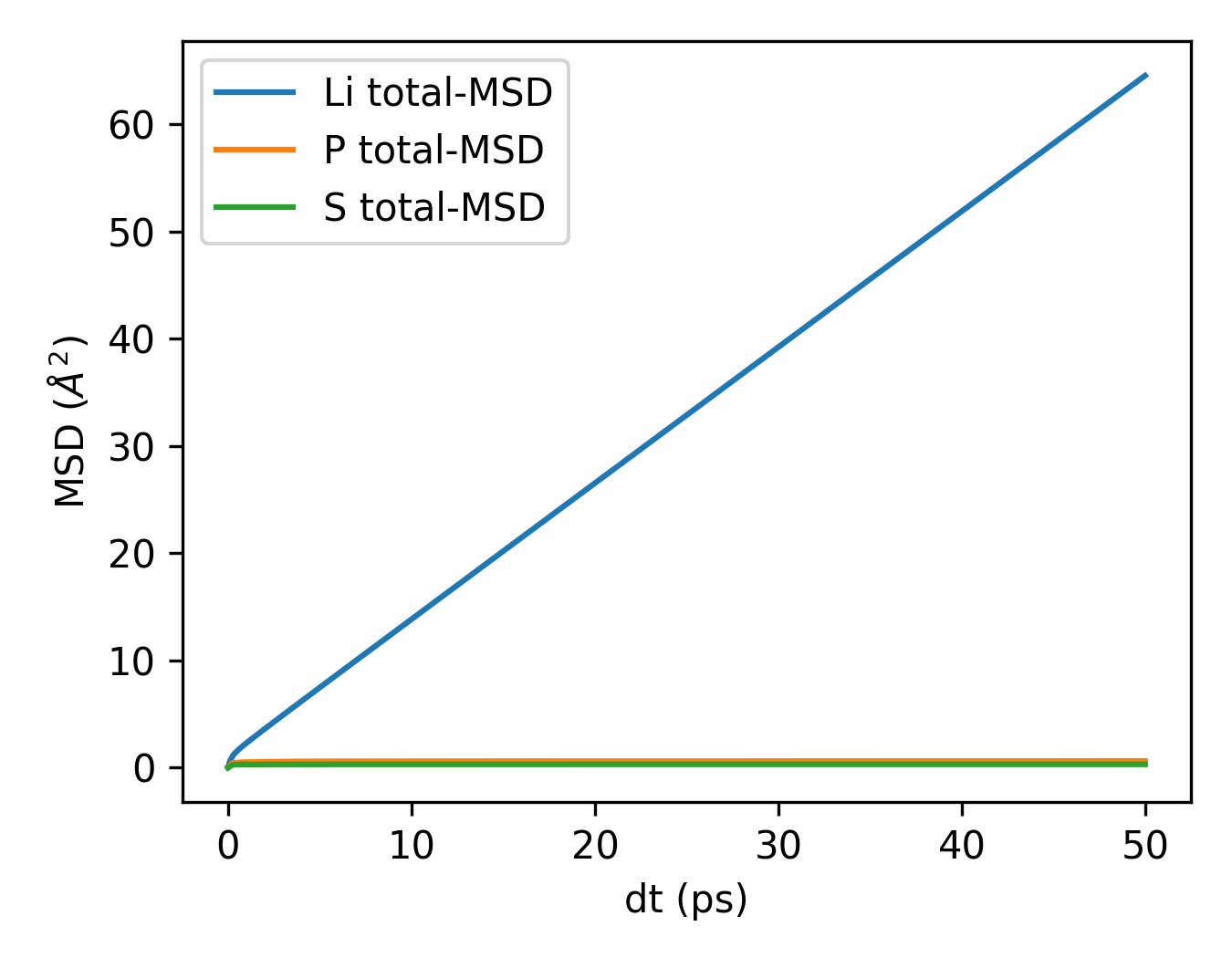
plt_msd_all.py
Plots MSD for all atomic species separately when using all_groups in GPUMD.
Input File: msd.out (computed with all_groups option)
Usage:
Requirements: Must use all_groups in the compute_msd command in run.in.
plt_msd_convergence_check.py
Checks convergence of MSD calculations across different time windows.
Input File: msd.out (computed with save_every option)
Usage:
Requirements: Use save_every in the compute_msd command.
Purpose: Verify MSD has converged sufficiently for accurate diffusion coefficient calculation.
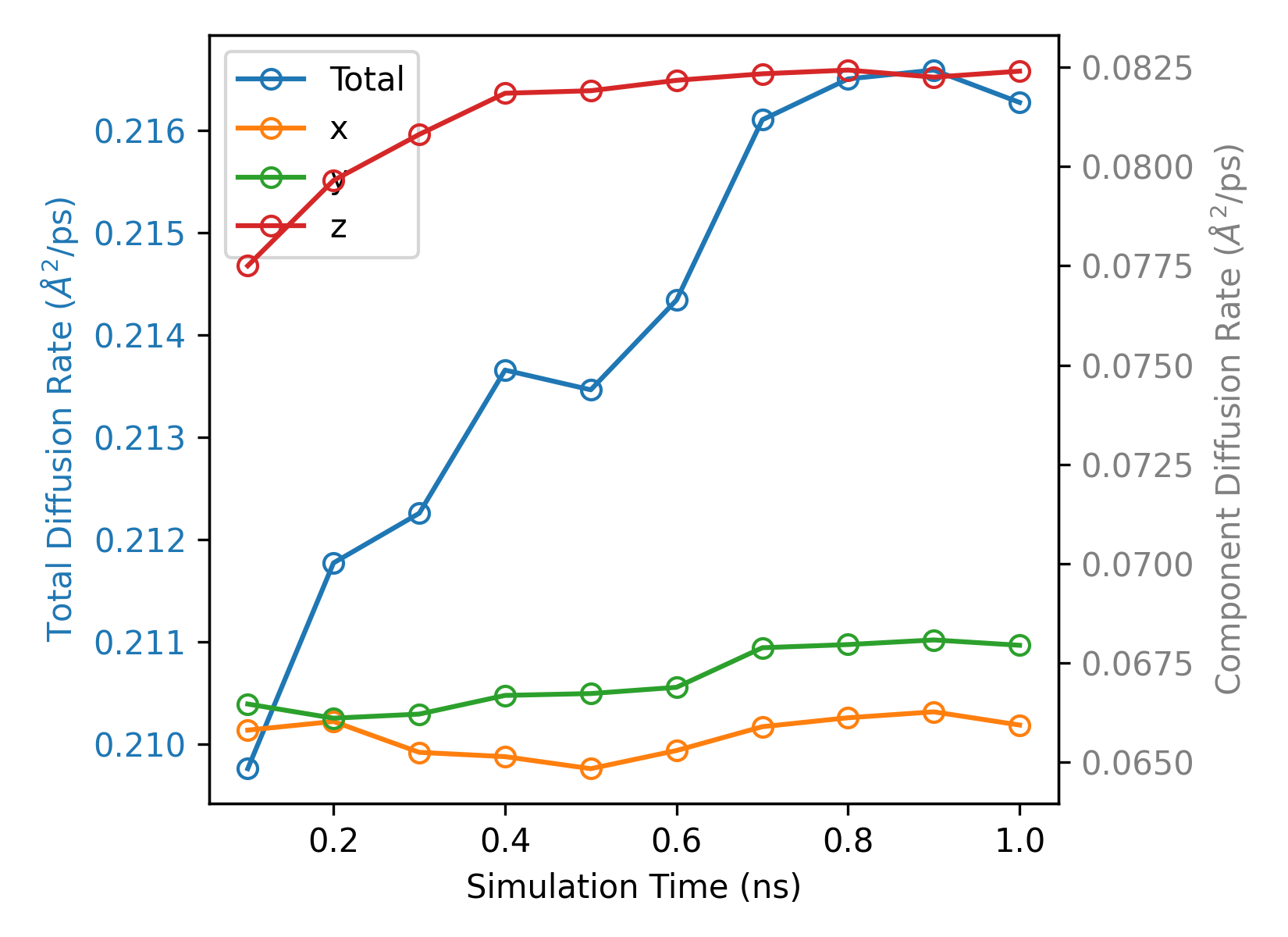
plt_sdc.py
Plots self-diffusion coefficient (SDC) vs time.
Input File: msd.out
Usage:
plt_arrhenius_d.py
Creates Arrhenius plot for diffusivity (log10 D vs 1000/T).
Input Files: *K/msd.out files
Usage:
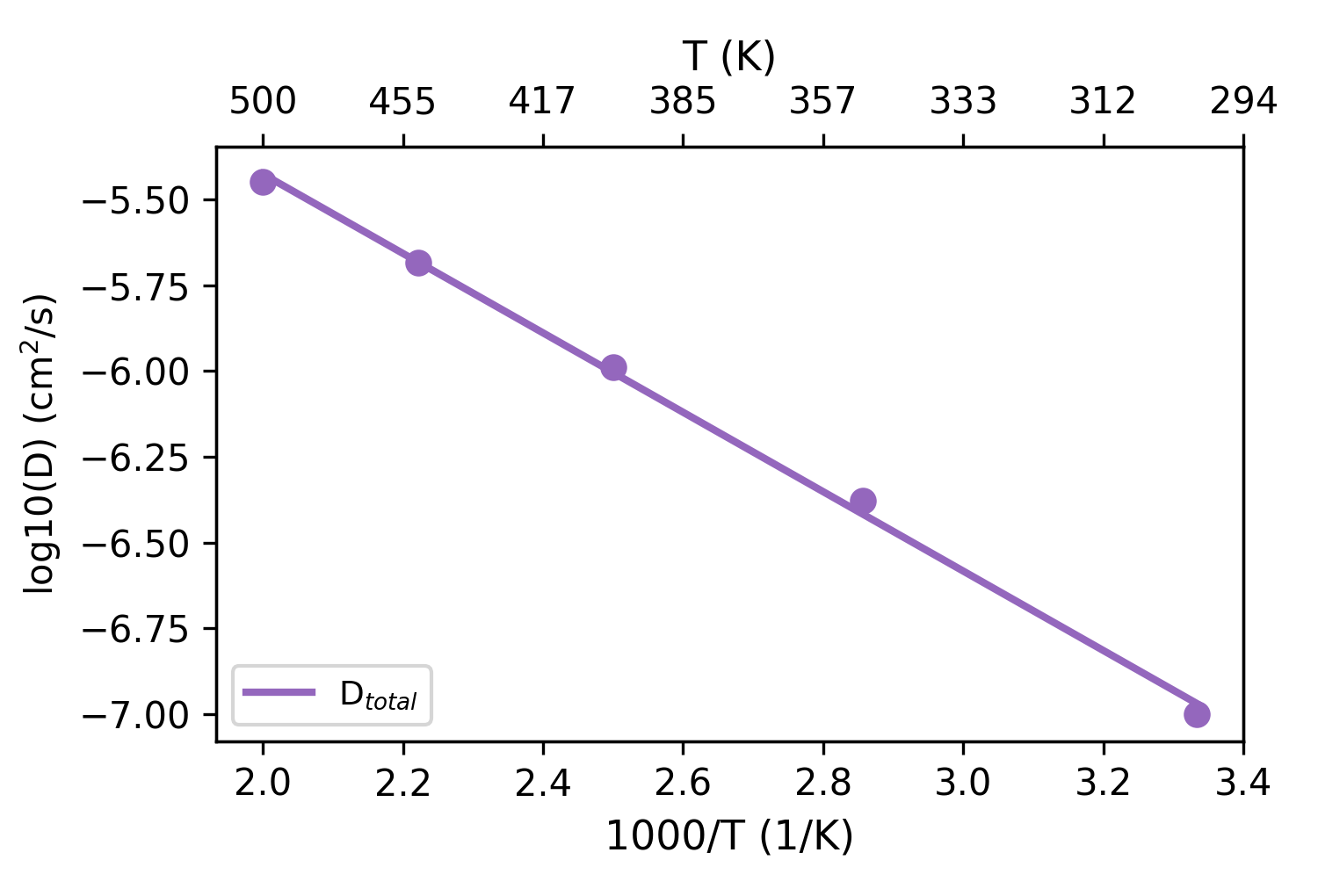
Output:
T: 300K, D_total: 1.001e-07 cm2/s
T: 350K, D_total: 4.184e-07 cm2/s
T: 400K, D_total: 1.027e-06 cm2/s
T: 450K, D_total: 2.069e-06 cm2/s
T: 500K, D_total: 3.563e-06 cm2/s
Ea: 0.230 eV
plt_arrhenius_sigma.py
Creates Arrhenius plot for ionic conductivity (ln(σ·T) vs 1000/T).
Input Files: *K/{model.xyz, run.in, thermo.out, msd.out}
Usage:
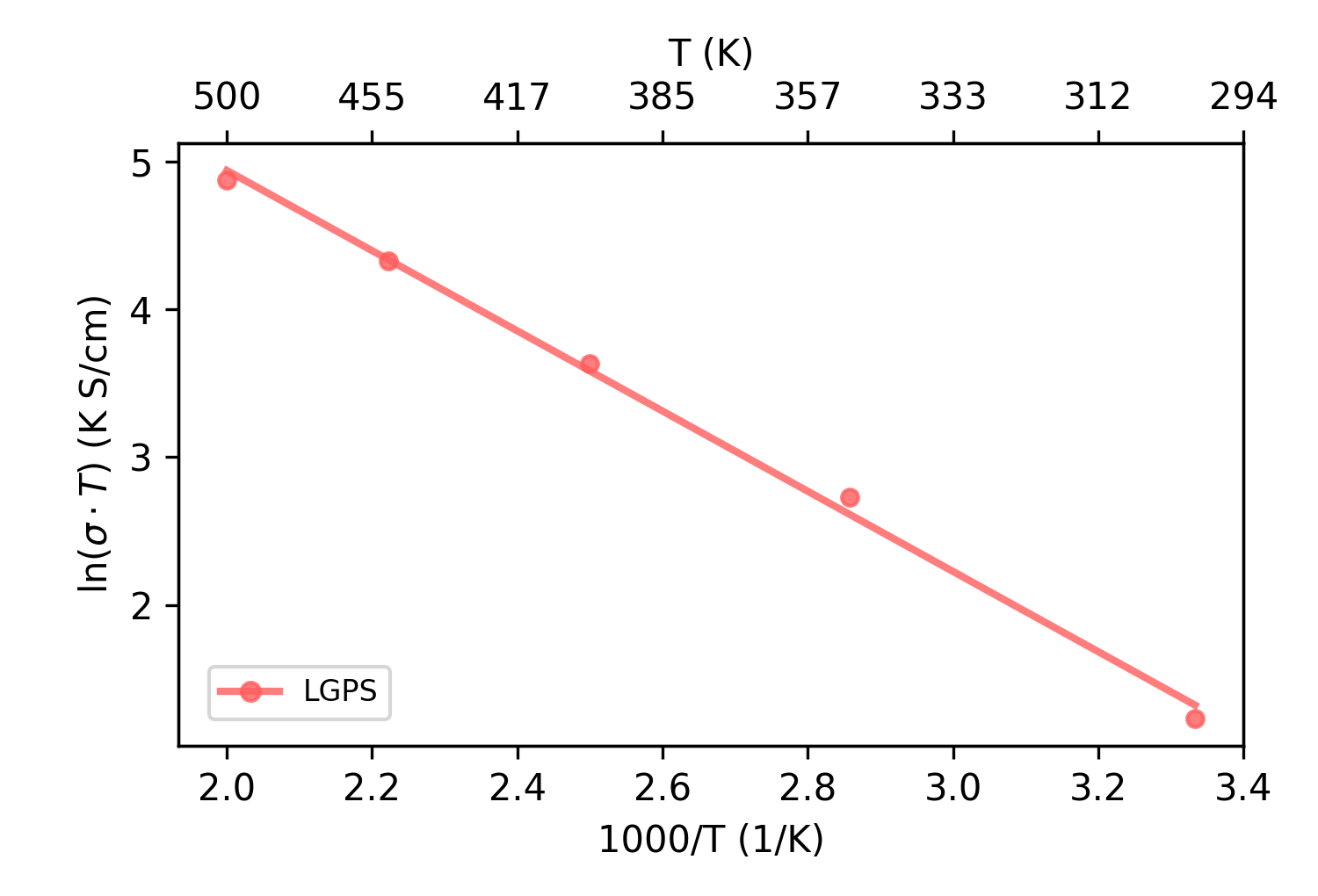
Output:
[Note] No run.in file found, assuming no replication
Conductivity Data:
T (K) Sigma (S/cm) Sigma·T (K·S/cm)
------------------------------------
300 1.141e-02 3.422e+00
350 4.377e-02 1.532e+01
400 9.470e-02 3.788e+01
450 1.688e-01 7.595e+01
500 2.625e-01 1.312e+02
------------------------------------
msd, Ea: 0.234 eV
at 300K, LGPS: Sigma = 1.250e-02 S/cm
plt_emd.py
Analyzes and plots thermal conductivity from equilibrium molecular dynamics (EMD).
Usage:
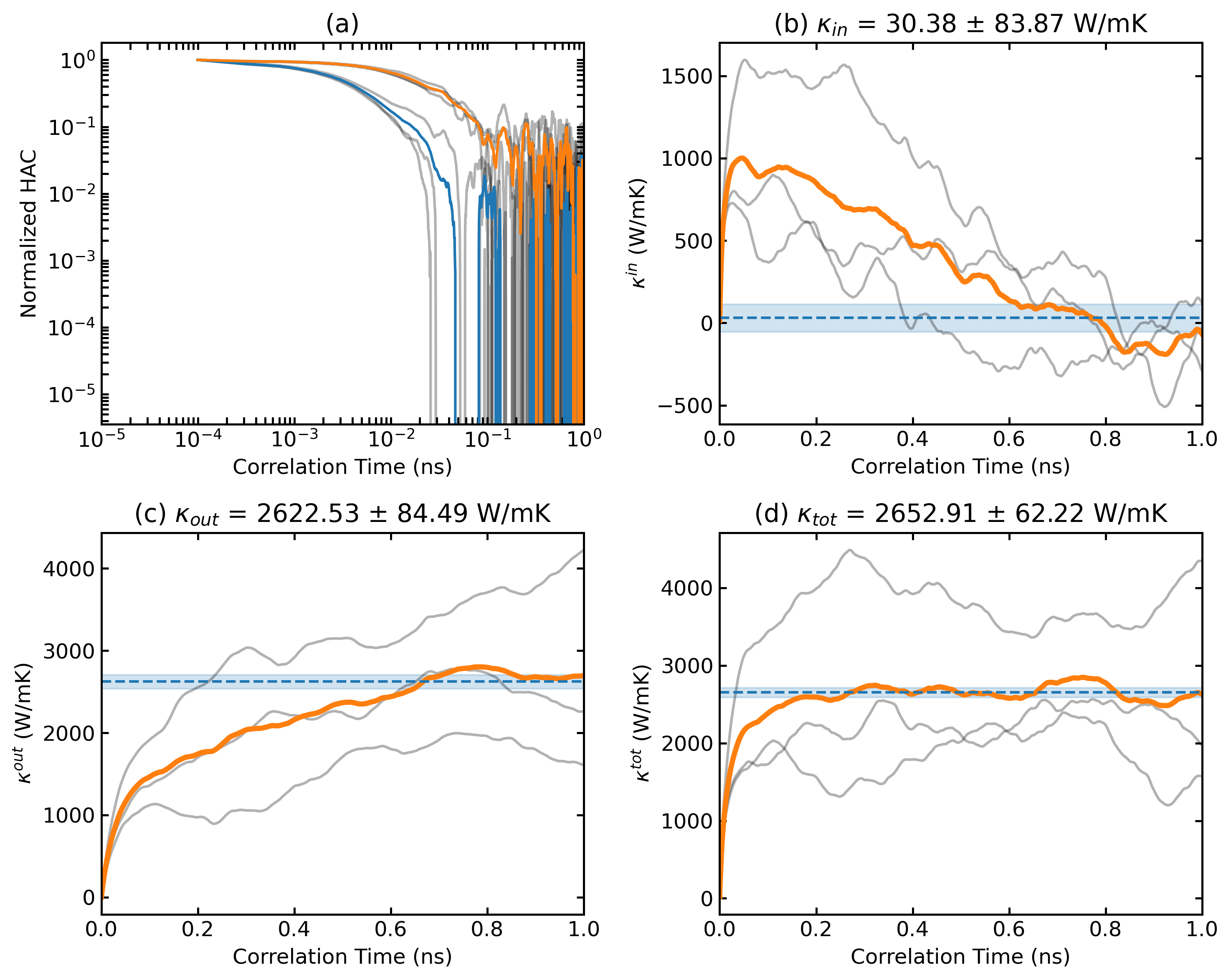
plt_nemd.py
Visualizes non-equilibrium molecular dynamics (NEMD) thermal transport properties.
Usage:
Params:
real_length : Real length of heat tranfer zone in nm (set to 'Auto', with auto-calculation)
scale_eff_size: Optional, Scale factor for effective cross-sectional area (default: 1)
• For 3D bulk systems: use 1
• For low-dimensional systems with vacuum layer: S_box / S_eff
- S_box: box area perpendicular to heat transfer direction
- S_eff: real or effective area of the system
cutoff_freq : Optional, Cutoff frequency for SHC calculation in THz (default: 60)
save : Optional, save the plot as 'nemd.png'
!!! Note !!! : If no SHC data, set [scale_eff_size] and [cutoff_freq] to any number as placeholders when using 'save'.
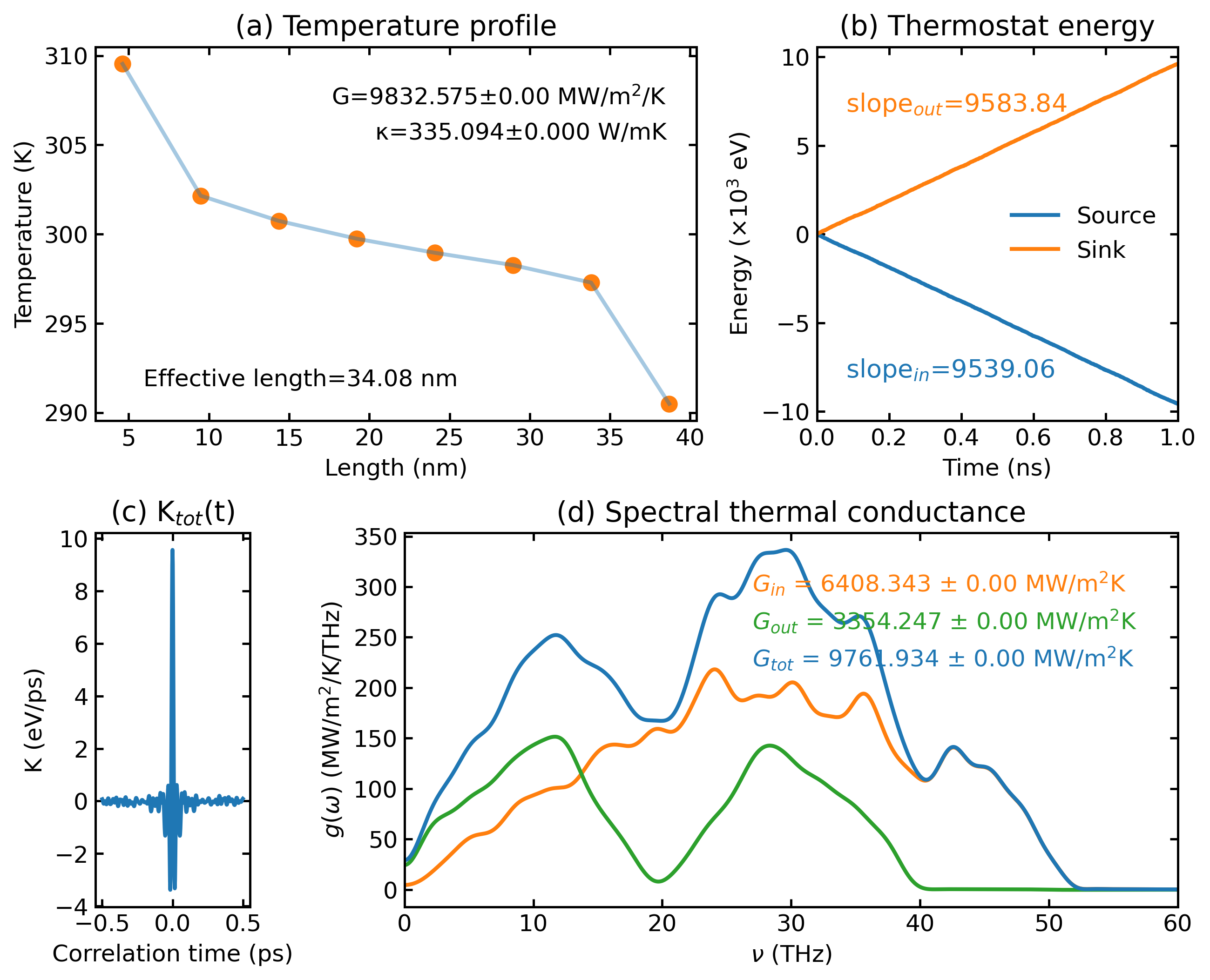
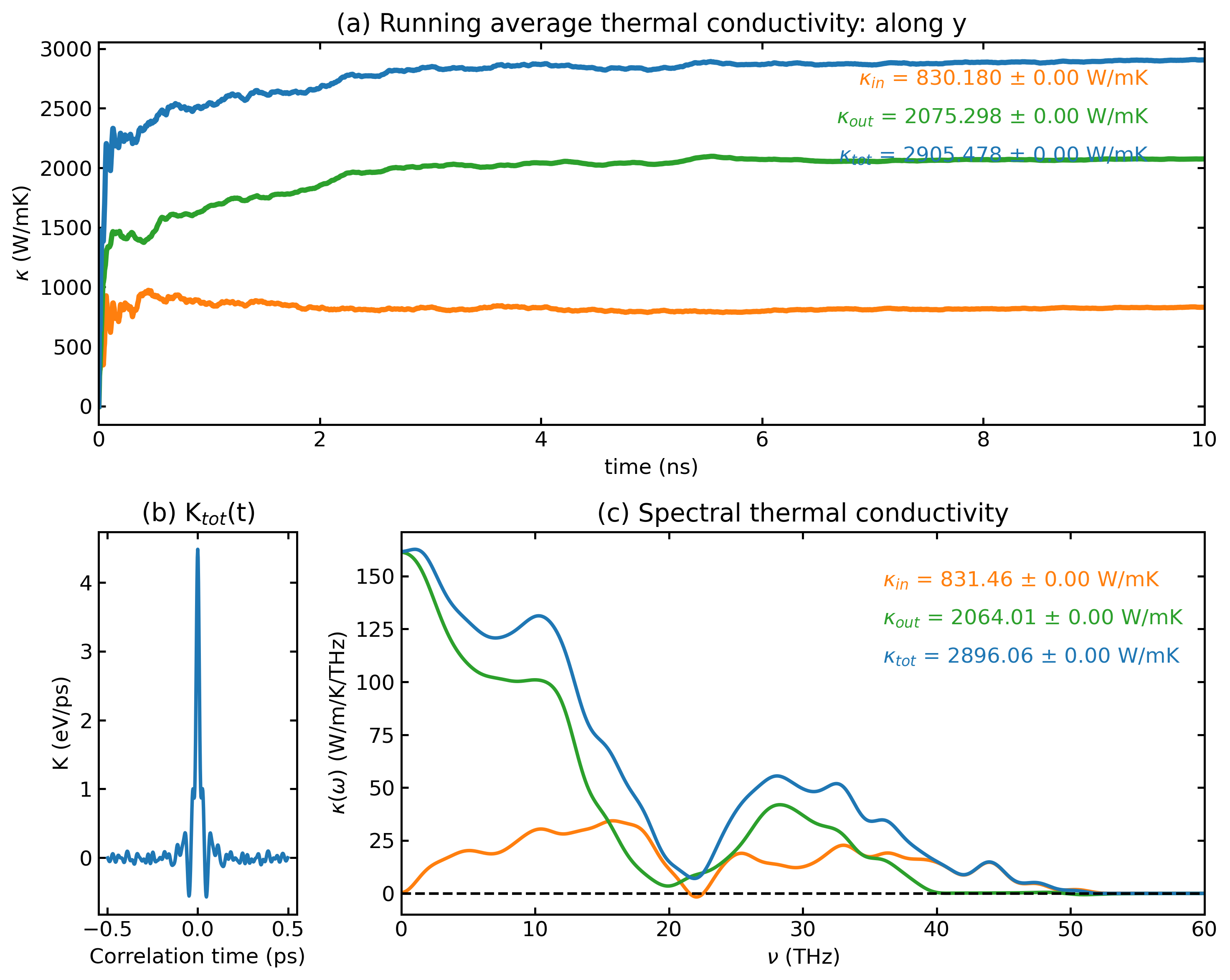
plt_hnemd.py
Plots homogeneous non-equilibrium molecular dynamics (HNEMD) results.
Usage:
Params:
scale_eff_size: Optional, Scale factor for effective cross-sectional area (default: 1)
• For 3D bulk systems: use 1
• For low-dimensional systems with vacuum layer: S_box / S_eff
- S_box: box area perpendicular to heat transfer direction
- S_eff: real or effective area of the system
cutoff_freq : Optional, Cutoff frequency for SHC calculation in THz (default: 60)
save : Optional, save the plot as 'hnemd.png'
!!! Note !!! : If no SHC data, set [scale_eff_size] and [cutoff_freq] to any number as placeholders when using 'save'.
plt_pdos.py
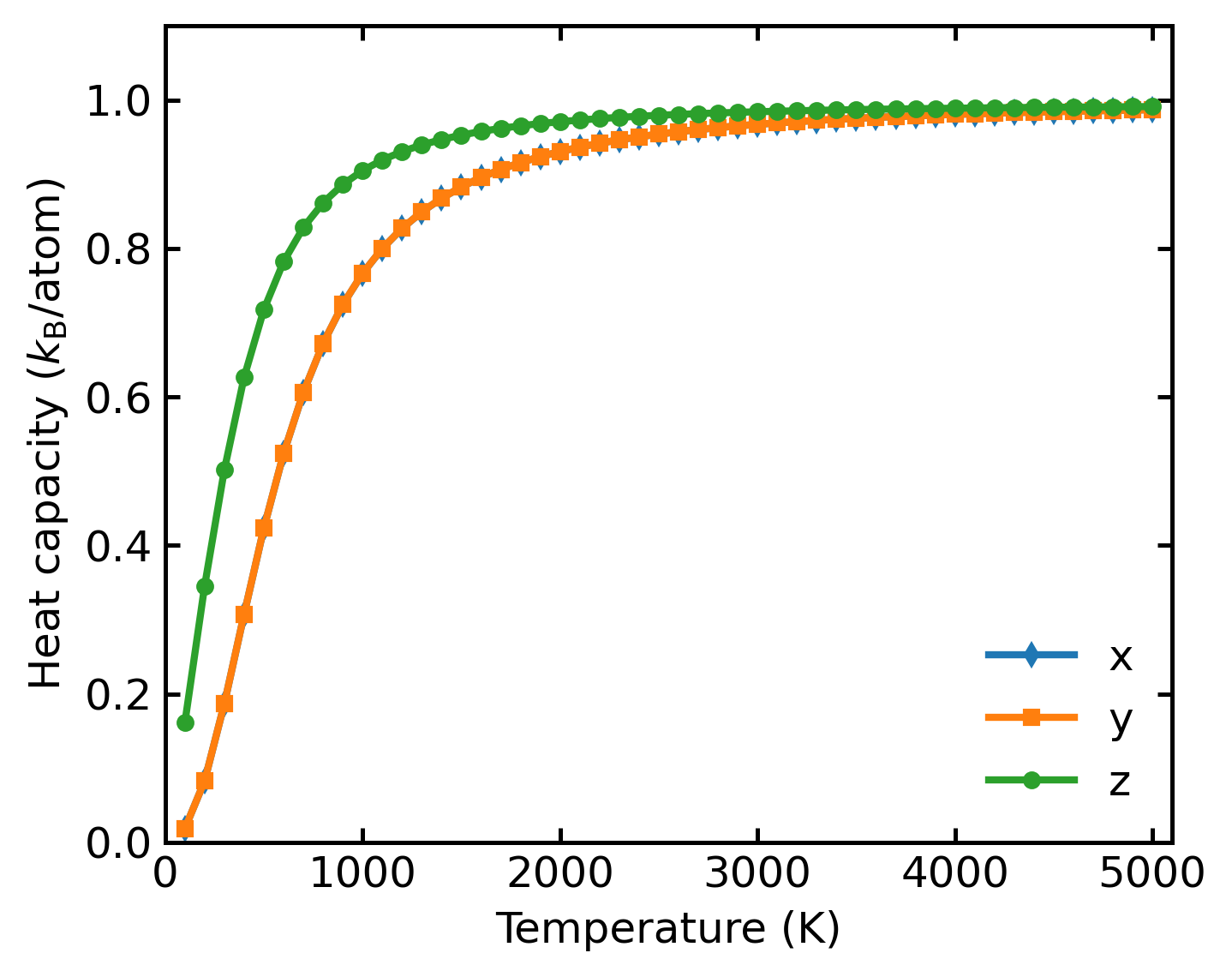
Calculates and Plots normalized VAC, PDOS, and Heat Capacity (Cv).
Usage:
Input Files: model.xyz, run.in, dos.out, mvac.out
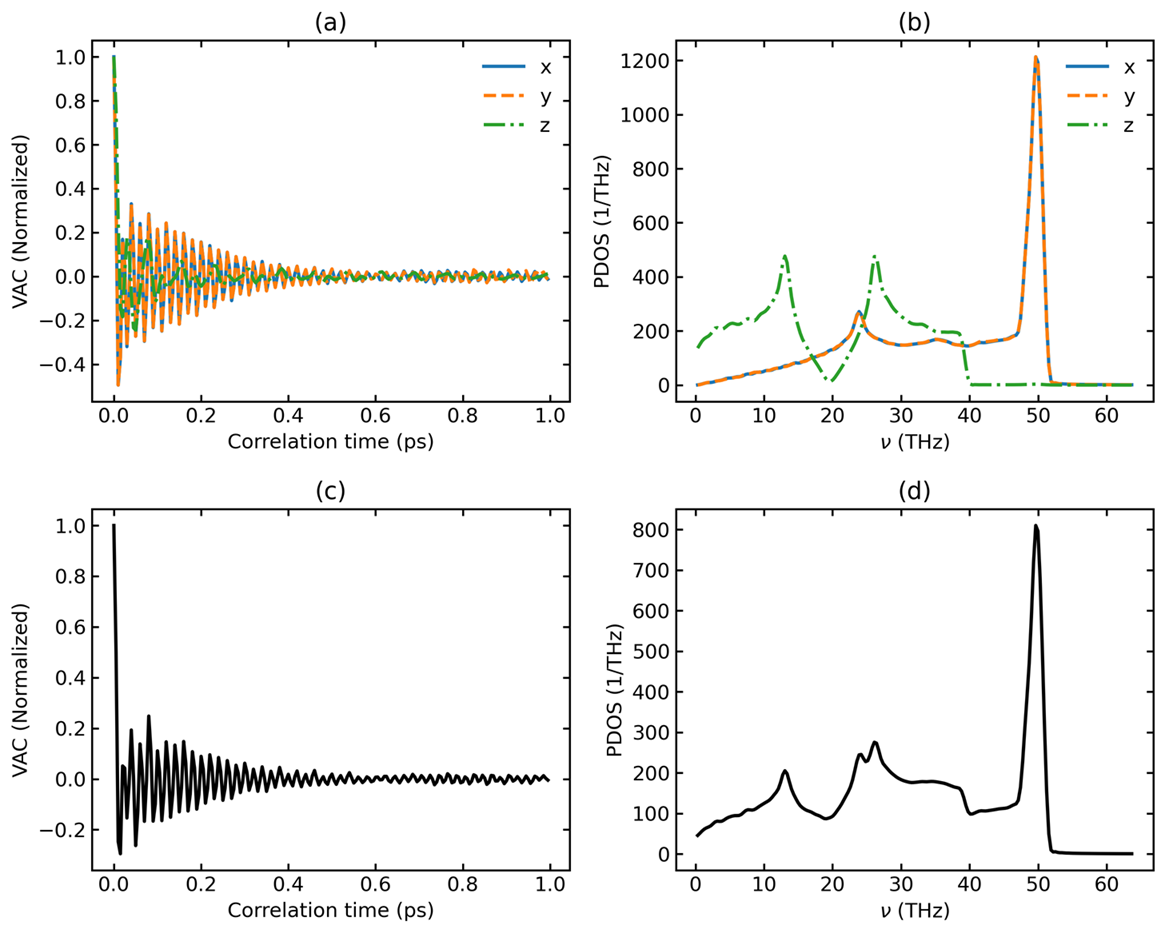
and heat capacity:
📐 Structural Analysis
Scripts for analyzing atomic structure and distributions.
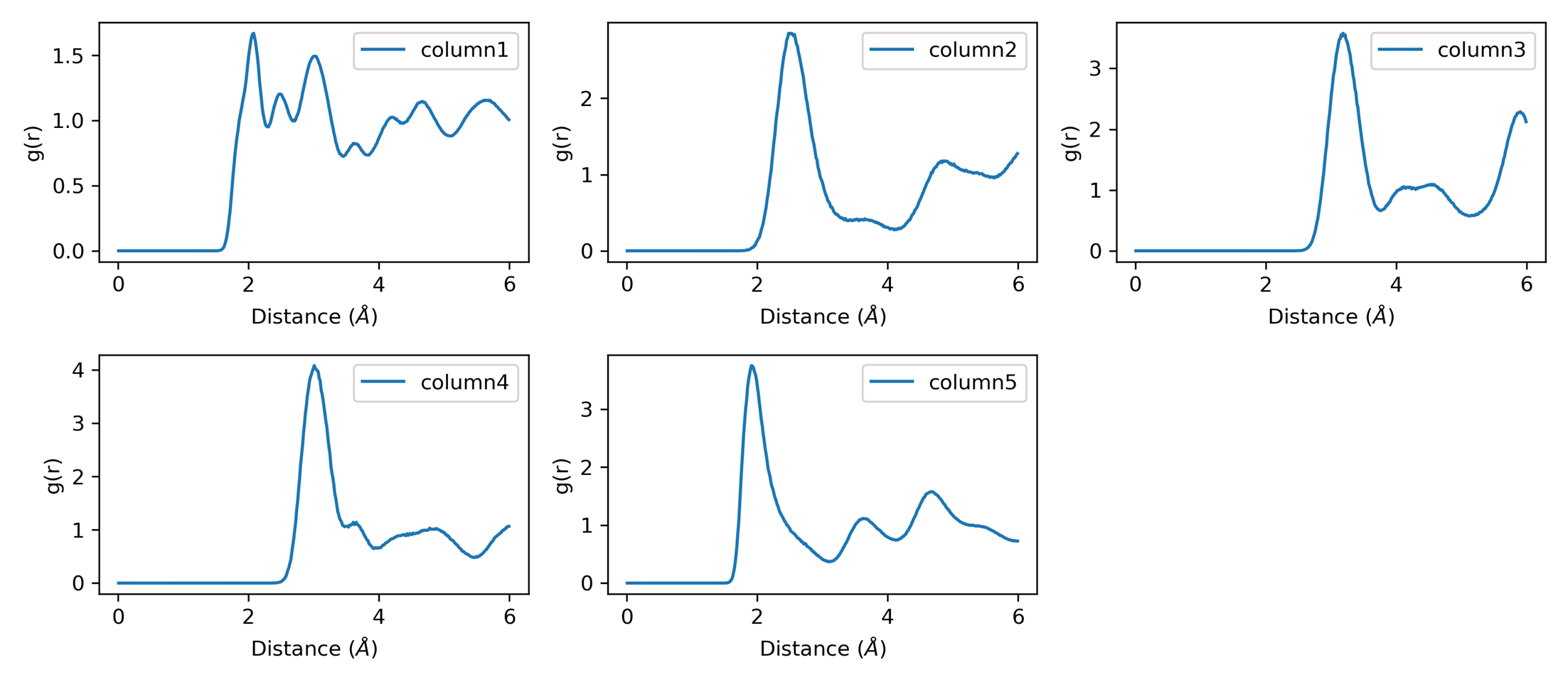
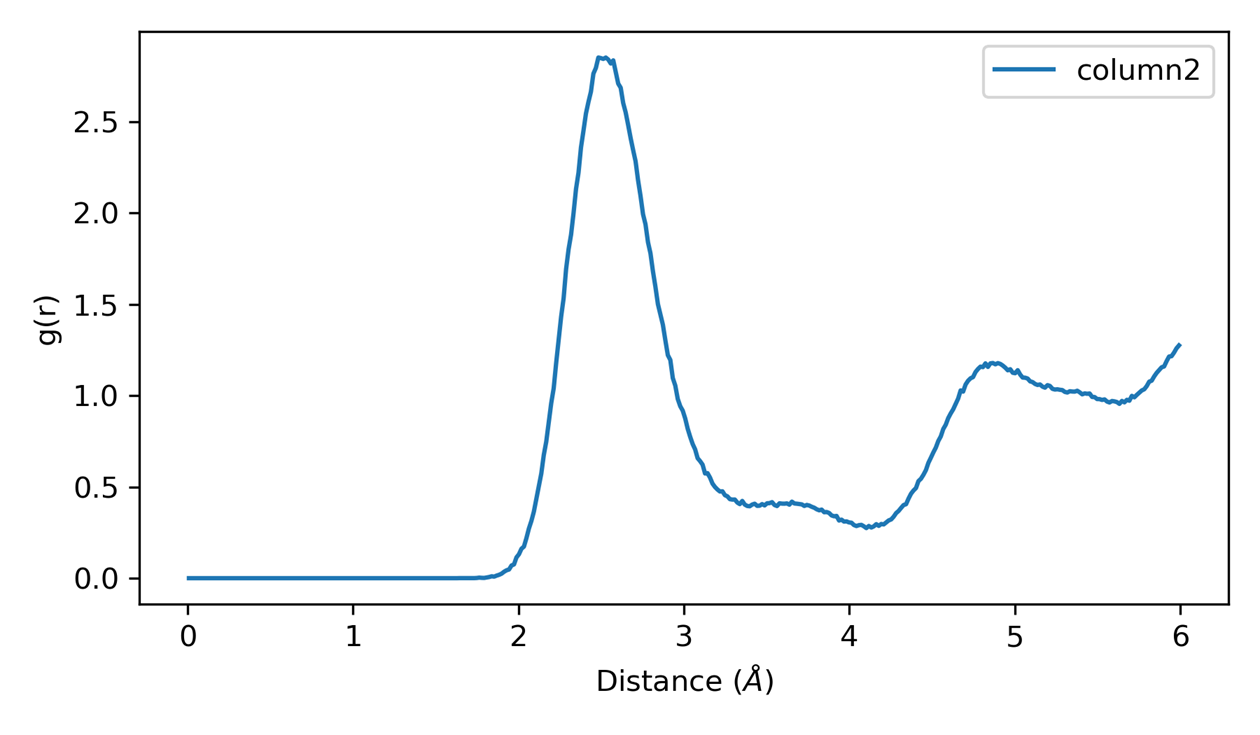
plt_rdf.py
Plots radial distribution function (RDF) showing pair correlations.
Input File: rdf.out
Usage:
# Plot all RDF pairs
gpumdkit.sh -plt rdf # Display all
# Plot specific column
gpumdkit.sh -plt rdf <column> # Display specific pair
Full RDF output:
Single pair RDF:
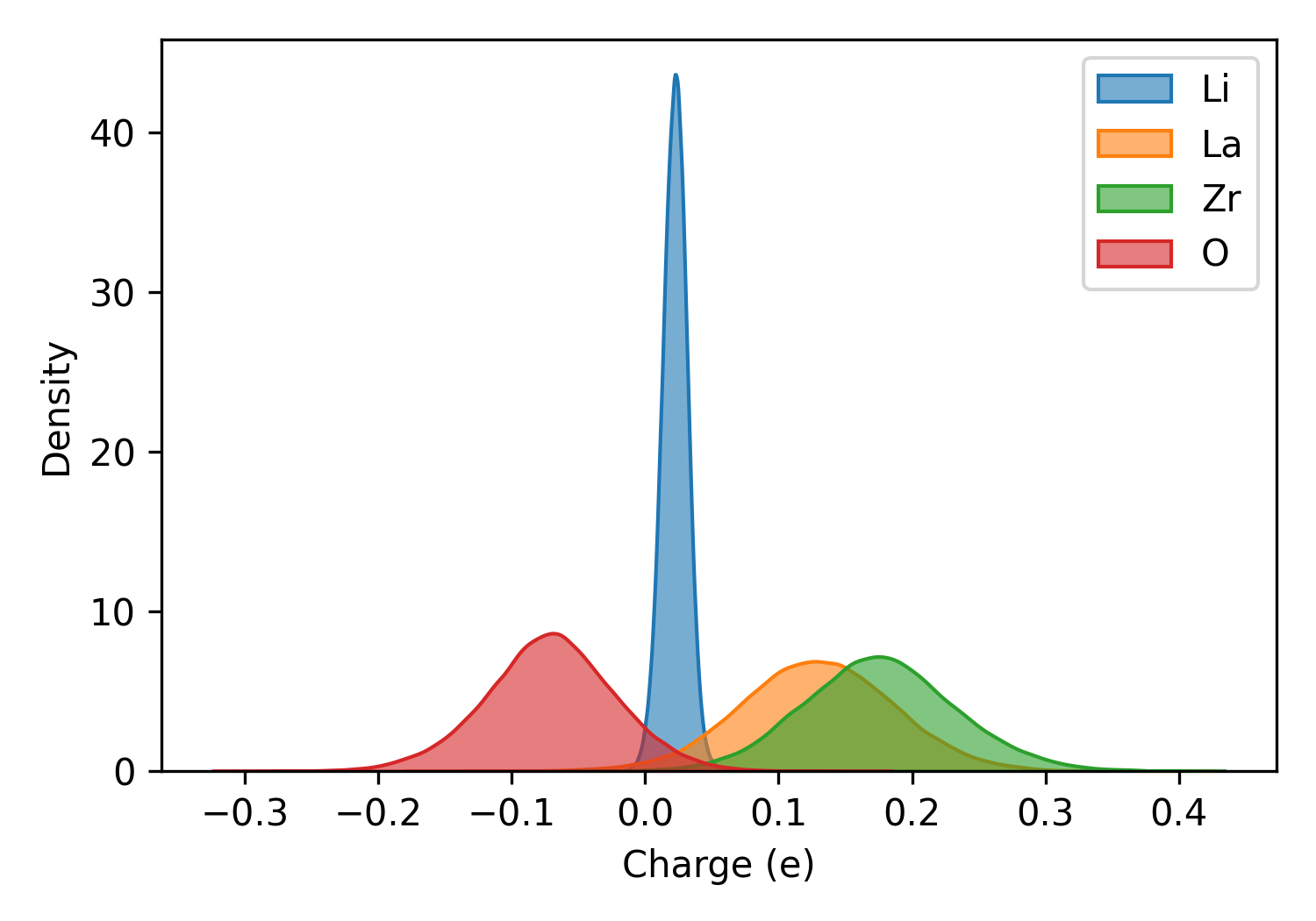
plt_charge.py
Plots charge distribution in charge_train.out from qNEP model.
Input File: charge_train.out
Usage:
Important: Ensure consistency between training set and charge output atom ordering. Use full batch training or run prediction step first.
🧬 Extra Analysis
Specialized scripts for some analysis.
plt_descriptors.py
Visualizes high-dimensional descriptors using dimensionality reduction (PCA/UMAP).
Input Files: descriptors.npy (generated by calc_descriptors.py)
Usage:
# First generate descriptors
gpumdkit.sh -calc des train.xyz descriptors.npy nep.txt Li
# Then plot
gpumdkit.sh -plt des <method>
Methods:
- pca - Principal Component Analysis
- umap - Uniform Manifold Approximation and Projection
You need some additional modifications to get the following plot, see the code here.
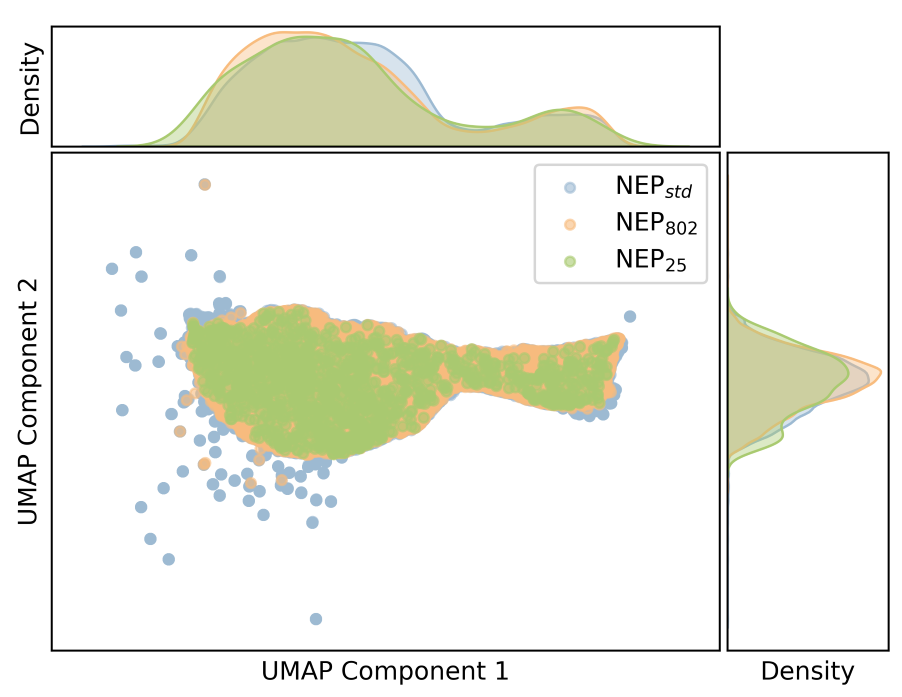
plt_descriptors_compare.py
Compares descriptors from multiple training sets or models.
plt_dimer.py
Plots dimer interaction curves. Two atoms are placed in a cubic box with the length of 30 angstrom. And then, the potential energy and the force will be calculated as a function of dimer distance.
Usage:
Example:
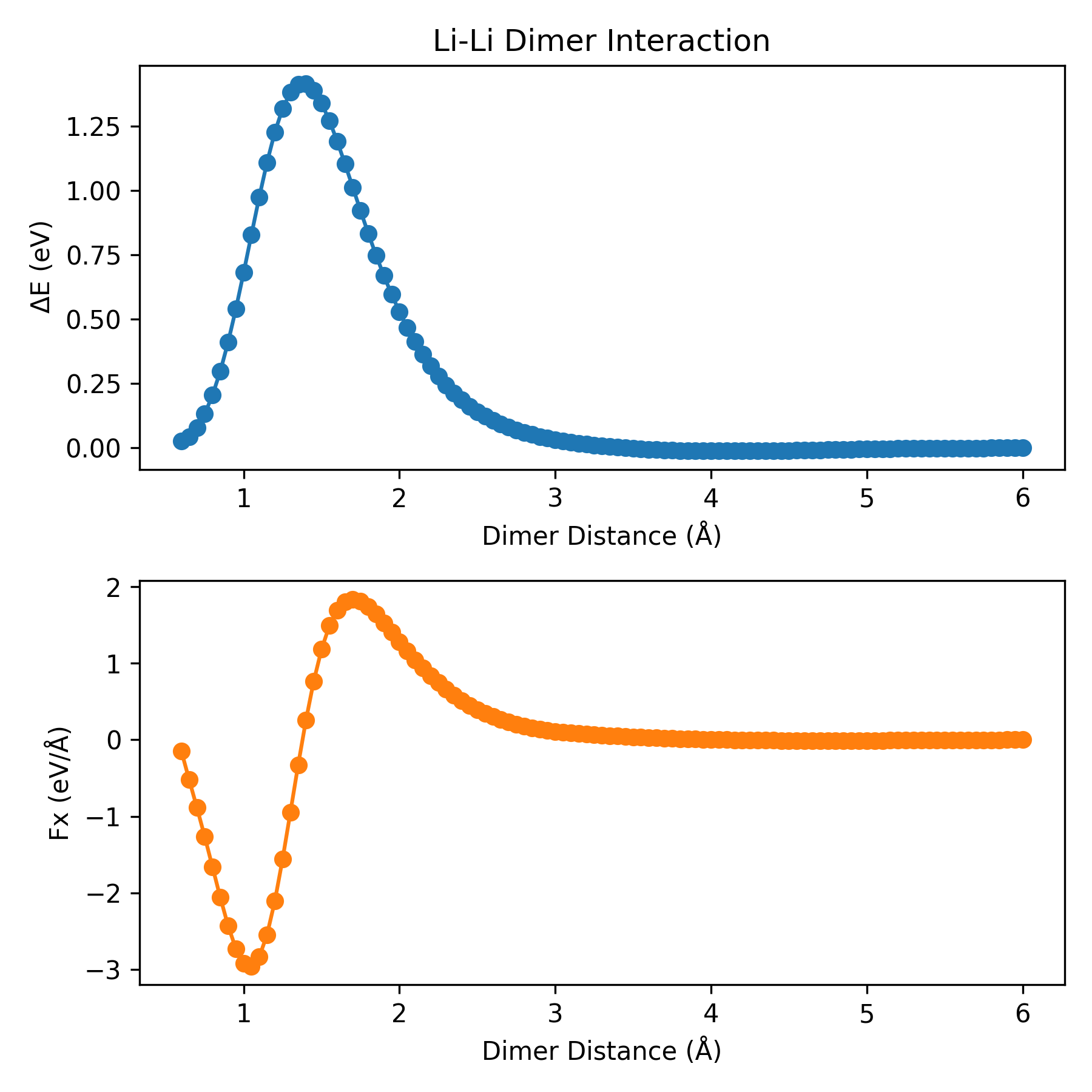
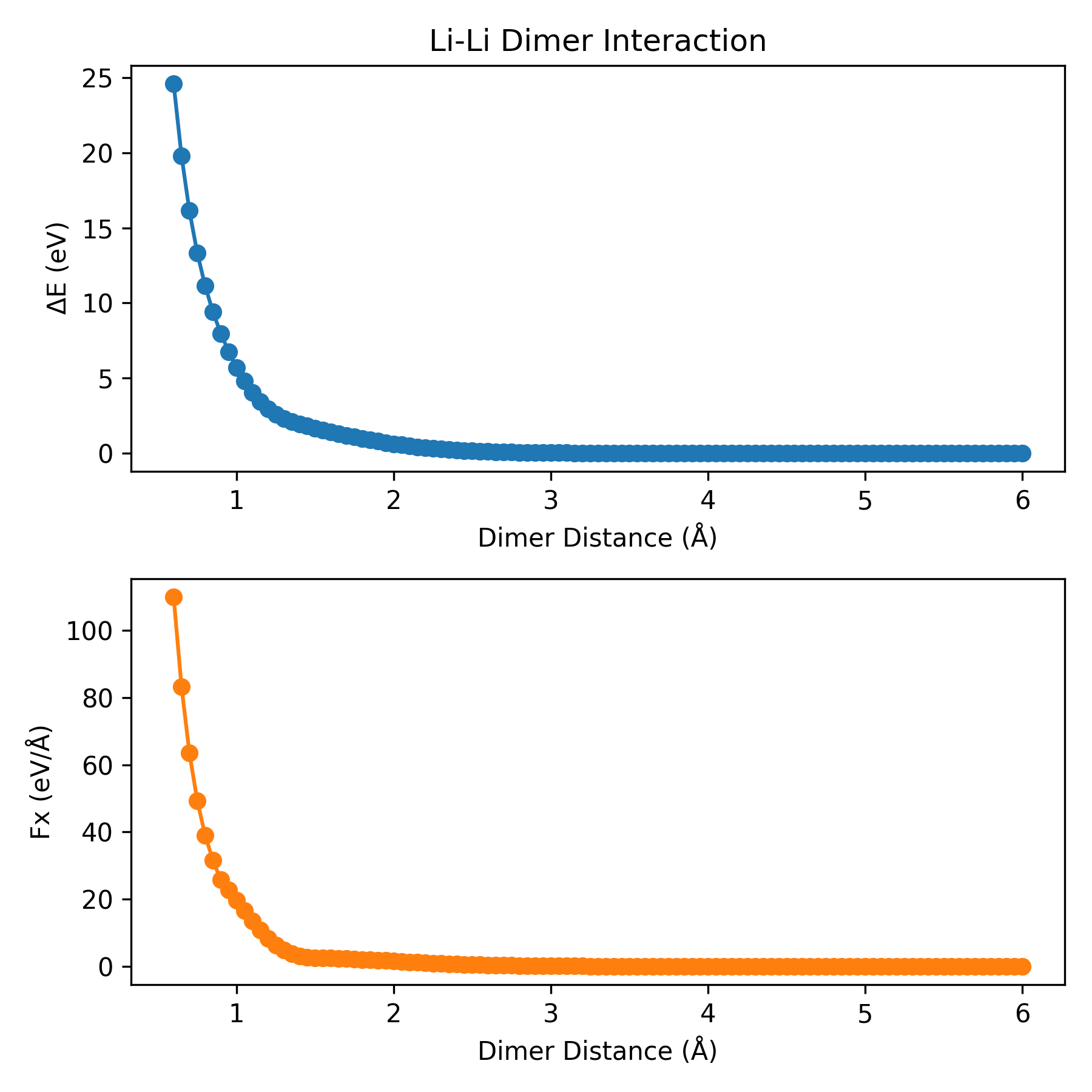
Reference: J. Chem. Inf. Model. 2026, 66, 3, 1406–1413
plt_doas.py
Plots density of atomistic states (DOAS) proposed by Wang et al..
Input Files: doas.out file calculated by gpumdkit.sh -calc doas
Usage:
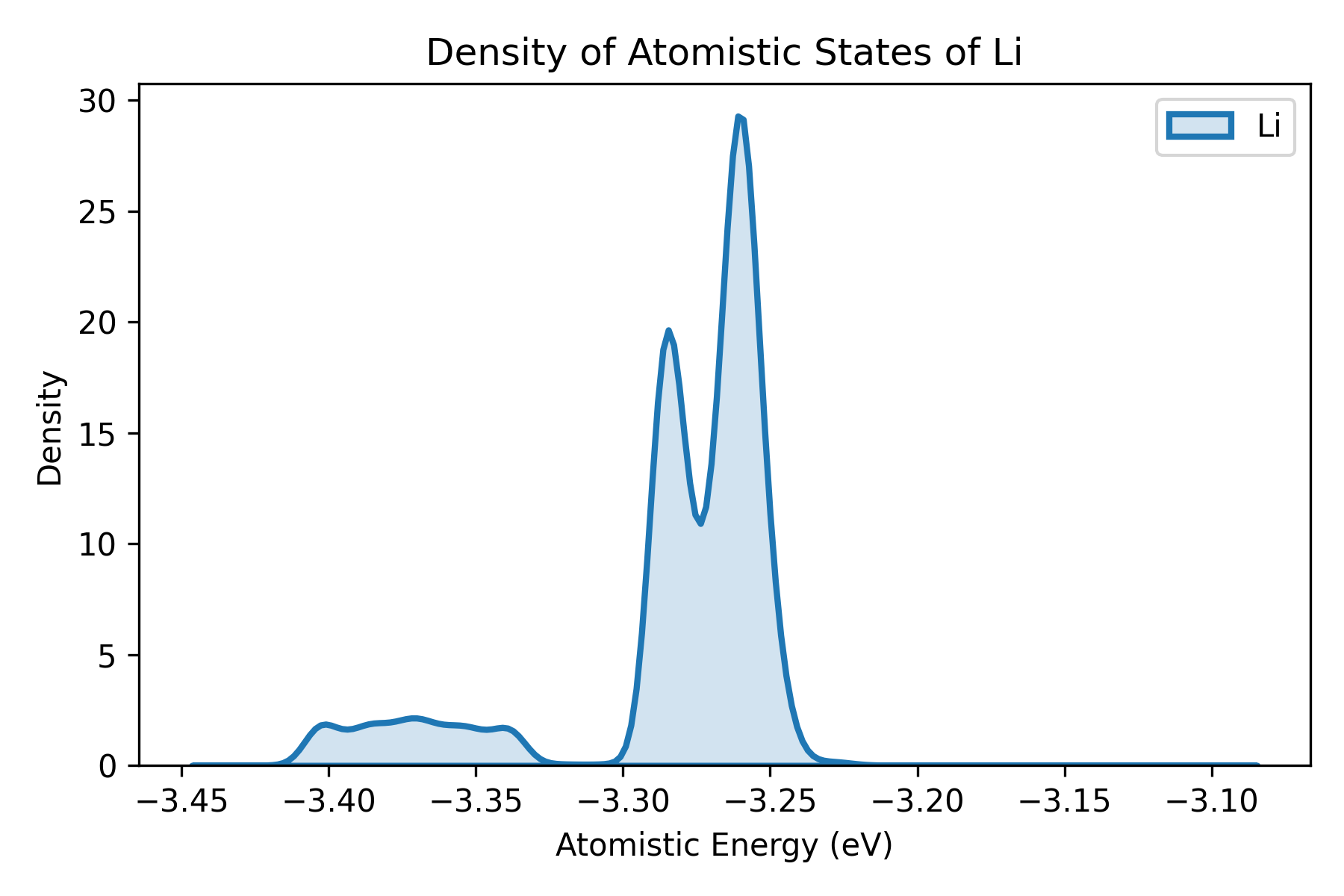
plt_net_force.py
Plots distribution of net forces on structures, useful for identifying problematic configurations.
Usage:
Reference: arXiv:2510.19774
Quick Reference Table
| Command | Input File(s) | Description |
|---|---|---|
thermo |
thermo.out |
Thermodynamic properties evolution |
train |
loss.out, *_train.out |
NEP training plot |
prediction |
*_test.out |
NEP prediction plot |
train_test |
*_train.out, *_test.out |
parity plots of train&test |
msd |
msd.out |
Mean square displacement |
sdc |
msd.out |
Self-diffusion coefficient |
rdf |
rdf.out |
Radial distribution function |
vac |
vac.out |
Velocity autocorrelation |
charge |
charge_train.out |
Charge distribution |
doas |
DOAS files | Density of atomistic states |
des |
descriptors.npy |
Descriptor visualization |
dimer |
Needs nep model | Dimer interaction curve |
force_errors |
force_train.out |
Force error metrics |
lr |
loss.out from gnep |
Learning rate |
restart |
nep.restart from nep |
Restart file parameters |
net_force |
train.xyz | Net force distribution |
emd |
EMD outputs | EMD thermal conductivity |
nemd |
NEMD outputs | NEMD thermal transport |
hnemd |
HNEMD outputs | HNEMD thermal transport |
arrhenius_d |
Multiple MSD | Arrhenius diffusivity |
arrhenius_sigma |
Conductivity data | Arrhenius conductivity |
Contributing
To add new plotting scripts:
- Create script following naming conventions
- Add documentation to this README
- Update gpumdkit.sh to integrate new plot type
See CONTRIBUTING.md for detailed guidelines.
Thank you for using GPUMDkit! If you have any questions, need further assistance, or want to contribute new plotting scripts, feel free to open an issue on our GitHub repository or contact Zihan YAN (yanzihan@westlake.edu.cn).