🔍 Analyzer Scripts
This directory (Scripts/analyzer/) contains analysis tools for structure files, simulation data, and dataset quality control. These scripts help validate, filter, and understand molecular dynamics data and training sets.
Script Location: Scripts/analyzer/
This section covers the analyzer tools in GPUMDkit (Interactive Mode - Option 5).
The analyzer scripts provide functionality for:
- Energy, force, and virial range analysis
- Minimum distance calculations (with/without periodic boundary conditions)
- Structure filtering by various criteria (distance, box size, property values)
- Composition analysis of multi-component systems
- Time estimation for GPUMD and NEP calculations
- Dataset quality checks and outlier detection
Interactive Mode
You'll see the following menu:
------------>>
501) Analyze composition of extxyz
502) Find outliers of extxyz
000) Return to the main menu
------------>>
for 501:
>-------------------------------------------------<
| This function calls the script in analyzer |
| Script: analyze_composition.py |
| Developer: Zihan YAN (yanzihan@westlake.edu.cn) |
>-------------------------------------------------<
Input <input.xyz> you want to analyze
Examp: train.xyz
------------>>
for 502:
>-------------------------------------------------<
| This function calls the script in analyzer |
| Script: find_outliers.py |
| Developer: Zihan YAN (yanzihan@westlake.edu.cn) |
>-------------------------------------------------<
Input the threshold of RMSE to identify outliers
---------------------------------------------------
Enter energy RMSE threshold (meV/atom): 1
Enter force RMSE threshold (meV/Å): 60
Enter stress RMSE threshold (GPa): 0.03
Follow the prompts to complete the function.
Command-Line Mode
gpumdkit.sh -range <file.xyz> <property>
gpumdkit.sh -min_dist <file.xyz>
gpumdkit.sh -min_dist_pbc <file.xyz>
gpumdkit.sh -filter_value <file.xyz> <property> <threshold>
gpumdkit.sh -filter_dist <file.xyz> <min_dist>
gpumdkit.sh -filter_box <file.xyz> <box_limit>
gpumdkit.sh -analyze_comp <file.xyz>
gpumdkit.sh -time gpumd
gpumdkit.sh -time nep
Scripts
analyze_composition.py
This script analyze the composition of your extxyz file.
Usage
Command-Line Mode Example
Output
Calling script by Zihan YAN
Code path: /d/Westlake/GPUMD/Gpumdkit/Scripts/analyzer/analyze_composition.py
Index Compositions N atoms Count
---------------------------------------------------
1 Li56O96Zr16La24 192 51
---------------------------------------------------
Enter index to export (e.g., '1,2', '2-3', 'all'), or press Enter to skip:
charge_balance_check.py
This script can be used to check the charge balance status of your extxyz file.
Usage
Command-Line Mode Example
Output
Calling script by Zihan YAN
Computing compositions: 100%|████████████████████████████████████████████████████████████████████| 51/51 [00:03<00:00, 13.03it/s]
Checking oxidation states: 100%|████████████████████████████████████████████████████████████████████| 1/1 [00:03<00:00, 3.67s/it]
Finally, you will get a balanced.xyz and indices.txt file.
energy_force_virial_analyzer.py
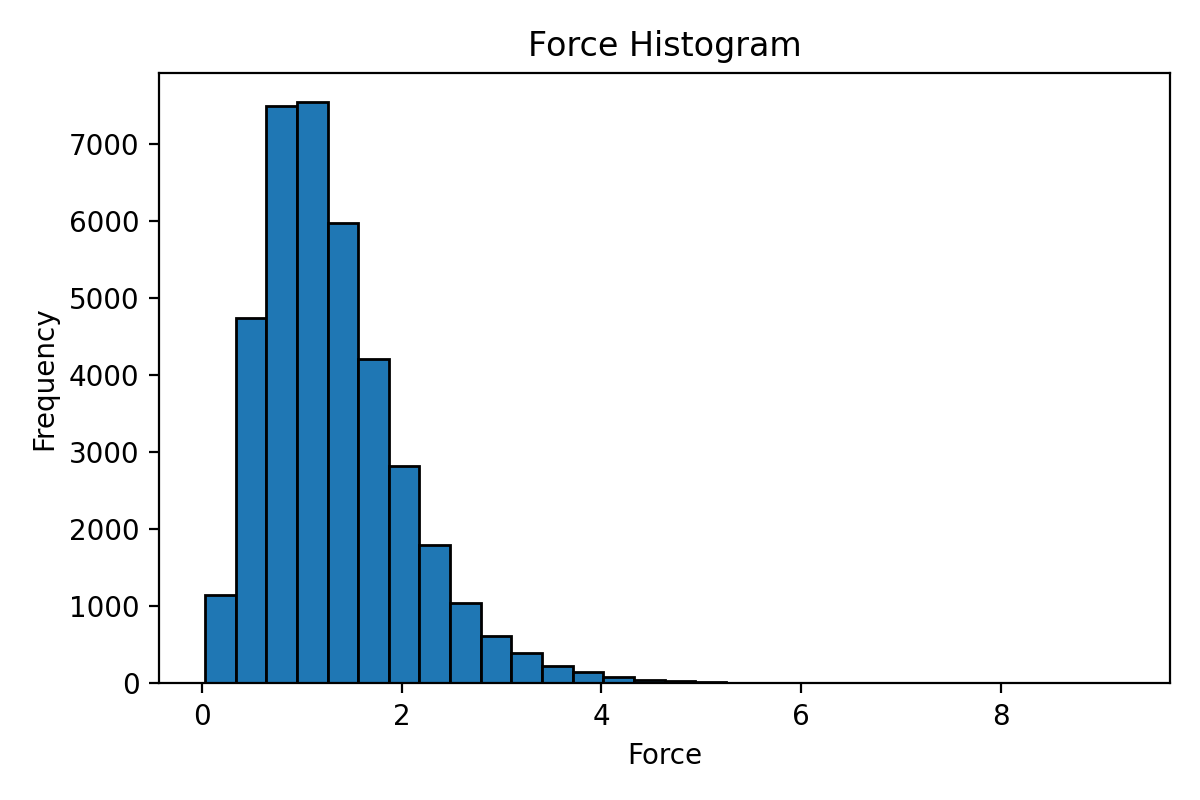
This script calculates and visualizes the range of properties (such as energy, forces, and virial) from the extxyz file.
Usage
The script requires at least two arguments:
- The filename of the
extxyzfile. - The name of the property to analyze (
energy,force, orvirial).
An optional third argument (hist) can be provided to generate a histogram plot of the values.
Example
Command-Line Mode Example
Output
If you add the [hist] option, it will calculate the range of forces and display a histogram:
filter_dist_range.py
This script is used to extract the structures with specified min_dist.
Usage
Example
Command-Line Mode Example
This means you need to extract the structures with the min_dist of Li-Li in the range of 1.8-2.0 in dump.xyz file. Finally, the filtered_Li_Li_1.8_2.0.xyz file will be generated.
get_min_dist.py
This script is used to calculate the min_dist of the structures.
Usage
Example
Command-Line Mode Example
Output
Calling script by Zihan YAN
Code path: /d/Westlake/GPUMD/Gpumdkit/Scripts/analyzer/get_min_dist.py
+---------------------------+
| PBC ignored for speed |
| use -min_dist_pbc for PBC |
+---------------------------+
Minimum interatomic distances:
+---------------------------+
| Atom Pair | Distance (Å) |
+---------------------------+
| Li-Li | 1.696 |
| Li-La | 2.498 |
| Li-Zr | 2.506 |
| Li-O | 1.587 |
| La-La | 3.463 |
| La-Zr | 3.243 |
| La-O | 2.043 |
| Zr-Zr | 5.060 |
| Zr-O | 1.867 |
| O-O | 2.480 |
+---------------------------+
Overall min_distance: 1.587 Å
NOTE: This script is fast because it does not take into account periodic boundary conditions (PBC), but in some cases it can be problematic.
get_min_dist_pbc.py
This script is used to calculate the min_dist of the structures considering the PBC.
Usage
Example
Command-Line Mode Example
Output
Calling script by Zihan YAN
Code path: /d/Westlake/GPUMD/Gpumdkit/Scripts/analyzer/get_min_dist_pbc.py
Minimum interatomic distances (with PBC):
+---------------------------+
| Atom Pair | Distance (Å) |
+---------------------------+
| Li-Li | 1.696 |
| Li-La | 2.498 |
| Li-Zr | 2.477 |
| Li-O | 1.587 |
| La-La | 3.463 |
| La-Zr | 3.210 |
| La-O | 2.043 |
| Zr-Zr | 5.060 |
| Zr-O | 1.867 |
| O-O | 2.355 |
+---------------------------+
Overall min_distance: 1.587 Å
find_outliers.py
This script is used to find outliers in training data based on RMSE thresholds for energy, force, and stress.
Usage
Interactive Mode
Input the function number:
5
------------>>
501) Analyze composition of extxyz
502) Find outliers of extxyz
000) Return to the main menu
------------>>
Input the function number:
502
>-------------------------------------------------<
| This function calls the script in analyzer |
| Script: find_outliers.py |
| Developer: Zihan YAN (yanzihan@westlake.edu.cn) |
>-------------------------------------------------<
Input the threshold of RMSE to identify outliers
---------------------------------------------------
Enter energy RMSE threshold (meV/atom): 1
Enter force RMSE threshold (meV/Å): 60
Enter stress RMSE threshold (GPa): 0.03
After that, you will get selected.xyz, remained.xyz, and slected_remained.png
filter_exyz_by_value.py
This script filter the structures by min_dist.
Usage
Example
Command-Line Mode Example
filter_exyz_by_box.py
This script filter the structures by box limit.
Usage
Example
Command-Line Mode Example
filter_exyz_by_value.py
This script filter the structures by specified value.
Usage
<extxyz_file>: The path to the inputextxyzfile.<property>: Filtering property:energy,force, orvirial<threshold>: Threshold value for filtering
Example
Command-Line Mode Example
This command will filter out the structure in train.xyz with a force greater than 20 eV/angstrom.
time_consuming_gpumd.sh
This script calculates the remaining time for GPUMD.
Usage
Command-Line Mode Example
Output
----------------- System Information ----------------
total frames: 1050000
-----------------------------------------------------
Current Frame Speed (steps/s) Total Time Time Left Estimated End
------------- ------------- ------------- ------------- -----------------
13000 499.86 0h 35m 0s 0h 34m 34s 2025-12-27 18:12:04
14000 199.93 1h 27m 31s 1h 26m 21s 2025-12-27 19:03:56
15000 199.94 1h 27m 31s 1h 26m 16s 2025-12-27 19:03:56
time_consuming_nep.sh
This script calculates the remaining time for nep.
Usage
Command-Line Mode Example
Output
+-----------------+-----------+-----------------+---------------------+
| Step | Time Diff | Time Left | Finish Time |
+-----------------+-----------+-----------------+---------------------+
| 6700 | 1 s | 0 h 15 m 33 s | 2025-10-23 15:34:11 |
| 6800 | 2 s | 0 h 31 m 4 s | 2025-10-23 15:49:44 |
| 6900 | 2 s | 0 h 31 m 2 s | 2025-10-23 15:49:44 |
| 7000 | 2 s | 0 h 31 m 0 s | 2025-10-23 15:49:44 |
| 7100 | 3 s | 0 h 46 m 27 s | 2025-10-23 16:05:14 |
Contributing
To add new analyzer scripts, see CONTRIBUTING.md for detailed guidelines.
Thank you for using GPUMDkit! If you have questions or need assistance with analyzer scripts, please open an issue on our GitHub repository or contact Zihan YAN (yanzihan@westlake.edu.cn).